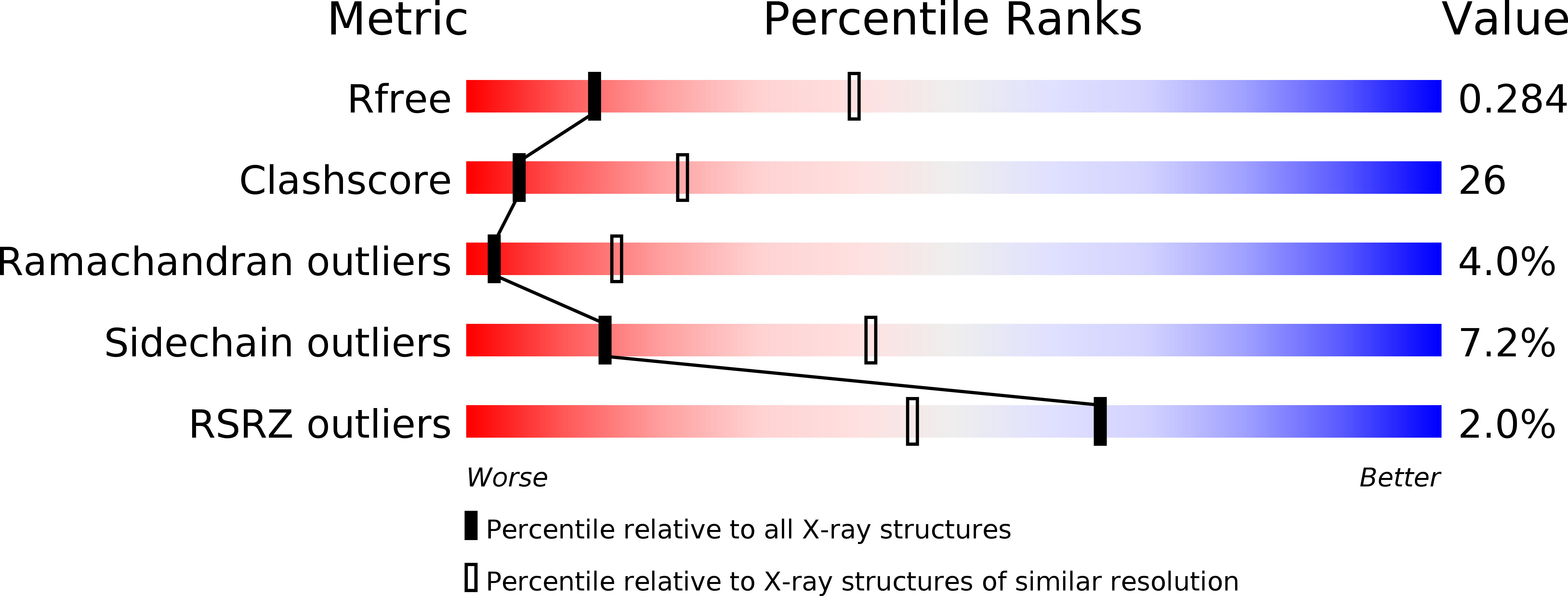
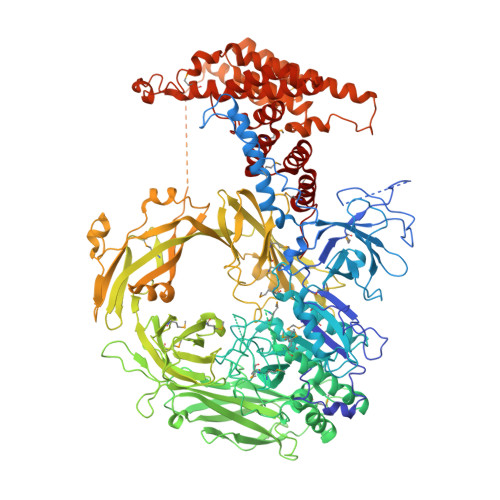
Hybrid molecular structure of the giant protease tripeptidyl peptidase II.
Chuang, C.K., Rockel, B., Seyit, G., Walian, P.J., Schonegge, A.M., Peters, J., Zwart, P.H., Baumeister, W., Jap, B.K.(2010) Nat Struct Mol Biol 17: 990-996
- PubMed: 20676100
- DOI: https://doi.org/10.1038/nsmb.1870
- Primary Citation of Related Structures:
3LXU - PubMed Abstract:
Tripeptidyl peptidase II (TPP II) is the largest known eukaryotic protease (6 MDa). It is believed to act downstream of the 26S proteasome, cleaving tripeptides from the N termini of longer peptides, and it is implicated in numerous cellular processes. Here we report the structure of Drosophila TPP II determined by a hybrid approach. We solved the structure of the dimer by X-ray crystallography and docked it into the three-dimensional map of the holocomplex, which we obtained by single-particle cryo-electron microscopy. The resulting structure reveals the compartmentalization of the active sites inside a system of chambers and suggests the existence of a molecular ruler determining the size of the cleavage products. Furthermore, the structure suggests a model for activation of TPP II involving the relocation of a flexible loop and a repositioning of the active-site serine, coupling it to holocomplex assembly and active-site sequestration.
Organizational Affiliation:
Life Sciences Division, Lawrence Berkeley National Laboratory, Berkeley, California, USA.