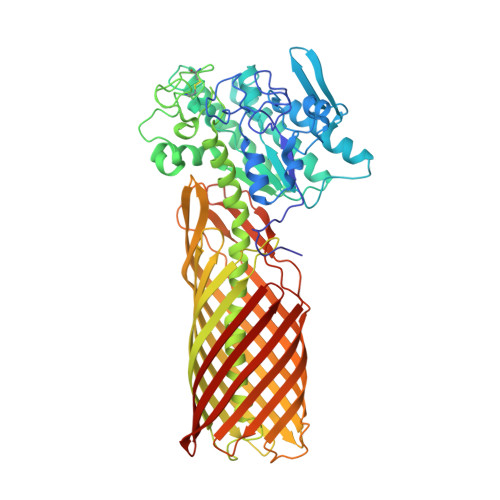
Crystal structure of a full-length autotransporter.
van den Berg, B.(2010) J Mol Biol 396: 627-633
- PubMed: 20060837
- DOI: https://doi.org/10.1016/j.jmb.2009.12.061
- Primary Citation of Related Structures:
3KVN - PubMed Abstract:
The autotransporter (AT) secretion mechanism is the most common mechanism for the secretion of virulence factors across the outer membrane (OM) from pathogenic Gram-negative bacteria. In addition, ATs have attracted biotechnological and biomedical interest for protein display on bacterial cell surfaces. Despite their importance, the mechanism by which passenger domains of ATs pass the OM is still unclear. The classical view is that the beta-barrel domain provides the conduit through which the unfolded passenger moves, with the energy provided by vectorial folding of the beta-strand-rich passenger on the extracellular side of the OM. We present here the first structure of a full-length AT, the esterase EstA from Pseudomonas aeruginosa, at a resolution of 2.5 A. EstA has a relatively narrow, 12-stranded beta-barrel that is covalently attached to the passenger domain via a long, curved helix that occupies the lumen of the beta-barrel. The passenger has a structure that is dramatically different from that of other known passengers, with a globular fold that is dominated by alpha-helices and loops. The arrangement of secondary-structure elements suggests that the passenger can fold sequentially, providing the driving force for passenger translocation. The esterase active-site residues are located at the apical surface of the passenger, at the entrance of a large hydrophobic pocket that contains a bound detergent molecule that likely mimics substrate. The EstA structure provides insight into AT mechanism and will facilitate the design of fusion proteins for cell surface display.
Organizational Affiliation:
Program in Molecular Medicine, University of Massachusetts Medical School, 373 Plantation Street, Worcester, MA 01605, USA. bert.vandenberg@umassmed.edu