Crystal structure of Putative sugar isomerase (YP_050048.1) from ERWINIA CAROTOVORA ATROSEPTICA SCRI1043 at 1.54 A resolution
Joint Center for Structural Genomics (JCSG)To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Xylose isomerase | 333 | Pectobacterium atrosepticum SCRI1043 | Mutation(s): 0 Gene Names: ECA1953 EC: 5.3.1.5 | 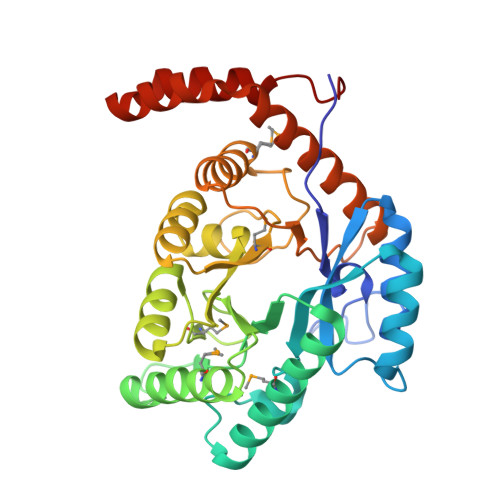 | |
UniProt | |||||
Find proteins for Q6D5T7 (Pectobacterium atrosepticum (strain SCRI 1043 / ATCC BAA-672)) Explore Q6D5T7 Go to UniProtKB: Q6D5T7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6D5T7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| EDO Query on EDO | AA [auth B] BA [auth B] I [auth A] J [auth A] K [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| FE Query on FE | C [auth A], D [auth A], O [auth B], P [auth B] | FE (III) ION Fe VTLYFUHAOXGGBS-UHFFFAOYSA-N |  | ||
| CL Query on CL | F [auth A], R [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| MG Query on MG | E [auth A], Q [auth B] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| UNL Query on UNL | G [auth A], H [auth A], S [auth B] | Unknown ligand JLVVSXFLKOJNIY-UHFFFAOYSA-N | |||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 79.653 | α = 90 |
| b = 79.653 | β = 90 |
| c = 194.406 | γ = 120 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| PHENIX | refinement |
| SHELX | phasing |
| MolProbity | model building |
| XSCALE | data scaling |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| SHELXD | phasing |
| autoSHARP | phasing |