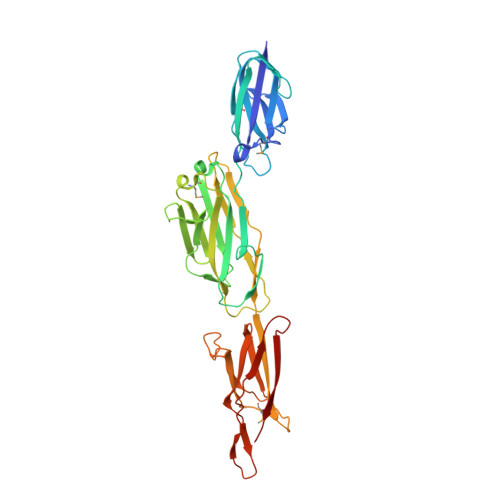
Intramolecular amide bonds stabilize pili on the surface of bacilli.
Budzik, J.M., Poor, C.B., Faull, K.F., Whitelegge, J.P., He, C., Schneewind, O.(2009) Proc Natl Acad Sci U S A 106: 19992-19997
- PubMed: 19903875
- DOI: https://doi.org/10.1073/pnas.0910887106
- Primary Citation of Related Structures:
3KPT - PubMed Abstract:
Gram-positive bacteria elaborate pili and do so without the participation of folding chaperones or disulfide bond catalysts. Sortases, enzymes that cut pilin precursors, form covalent bonds that link pilin subunits and assemble pili on the bacterial surface. We determined the x-ray structure of BcpA, the major pilin subunit of Bacillus cereus. The BcpA precursor encompasses 2 Ig folds (CNA(2) and CNA(3)) and one jelly-roll domain (XNA) each of which synthesizes a single intramolecular amide bond. A fourth amide bond, derived from the Ig fold of CNA(1), is formed only after pilin subunits have been incorporated into pili. We report that the domains of pilin precursors have evolved to synthesize a discrete sequence of intramolecular amide bonds, thereby conferring structural stability and protease resistance to pili.
Organizational Affiliation:
Departments of Microbiology and Chemistry, University of Chicago, Chicago, IL 60637, USA.