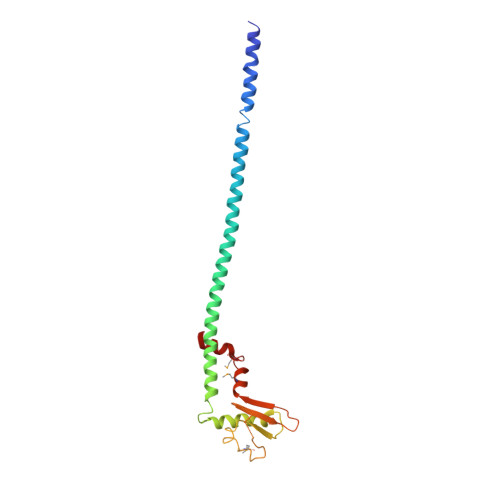
The structure of the peripheral stalk of Thermus thermophilus H(+)-ATPase/synthase.
Lee, L.K., Stewart, A.G., Donohoe, M., Bernal, R.A., Stock, D.(2010) Nat Struct Mol Biol 17: 373-378
- PubMed: 20173764
- DOI: https://doi.org/10.1038/nsmb.1761
- Primary Citation of Related Structures:
3K5B - PubMed Abstract:
Proton-translocating ATPases are ubiquitous protein complexes that couple ATP catalysis with proton translocation via a rotary catalytic mechanism. The peripheral stalks are essential components that counteract torque generated from proton translocation during ATP synthesis or from ATP hydrolysis during proton pumping. Despite their essential role, the peripheral stalks are the least conserved component of the complexes, differing substantially between subtypes in composition and stoichiometry. We have determined the crystal structure of the peripheral stalk of the A-type ATPase/synthase from Thermus thermophilus consisting of subunits E and G. The structure contains a heterodimeric right-handed coiled coil, a protein fold never observed before. We have fitted this structure into the 23 A resolution EM density of the intact A-ATPase complex, revealing the precise location of the peripheral stalk and new implications for the function and assembly of proton-translocating ATPases.
Organizational Affiliation:
Structural and Computational Biology Division, The Victor Chang Cardiac Research Institute, Darlinghurst, Australia.