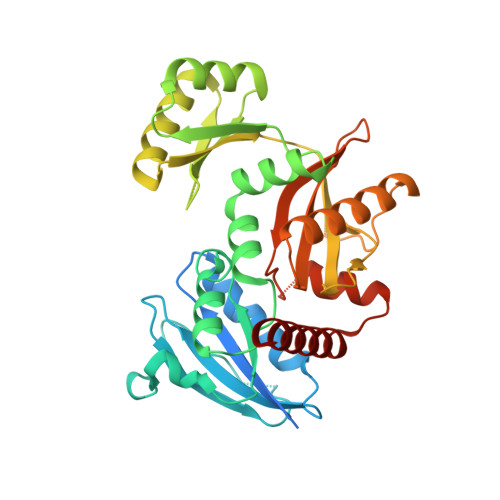
Crystal structure of the Apo form of D-Alanine:D-Alanine ligase (DDl) from Streptococcus mutans.
Lu, Y., Xu, H., Zhao, X.(2010) Protein Pept Lett 17: 1053-1057
- PubMed: 20522004
- DOI: https://doi.org/10.2174/092986610791498858
- Primary Citation of Related Structures:
3K3P - PubMed Abstract:
D-Alanine:D-Alanine ligase (DDl) catalyzes the formation of D-Alanine:D-Alanine dipeptide and is an essential enzyme in bacterial cell wall biosynthesis.. This enzyme does not have a human ortholog, making it an attractive target for developing new antibiotic drugs. We determined the crystal structure at 2.23 A resolution of DDl from Streptococcus mutans (SmDDl), the principal aetiological agent of human dental caries. This structure reveals that SmDDl is a dimer and has a disordered omega-loop region.
Organizational Affiliation:
West China Hospital Nanomedicine Laboratory, Center for Regenerative Medicine and Institute for Nanobiomedical Technology and Membrane Biology, West China Hospital Sichuan University, Chengdu 610065, Sichuan, China.