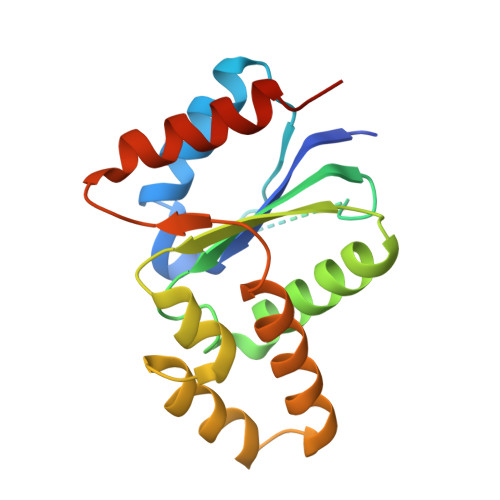
The crystal structure of a TIR domain from Arabidopsis thaliana reveals a conserved helical region unique to plants.
Chan, S.L., Mukasa, T., Santelli, E., Low, L.Y., Pascual, J.(2009) Protein Sci 19: 155-161
- PubMed: 19845004
- DOI: https://doi.org/10.1002/pro.275
- Primary Citation of Related Structures:
3JRN - PubMed Abstract:
Plants use a highly evolved immune system to exhibit defense response against microbial infections. The plant TIR domain, together with the nucleotide-binding (NB) domain and/or a LRR region, forms a type of molecule, named resistance (R) proteins, that interact with microbial effector proteins and elicit hypersensitive responses against infection. Here, we report the first crystal structure of a plant TIR domain from Arabidopsis thaliana (AtTIR) solved at a resolution of 2.0 A. The structure consists of five beta-strands forming a parallel beta-sheet at the core of the protein. The beta-strands are connected by a series of alpha-helices and the overall fold mimics closely that of other mammalian and bacterial TIR domains. However, the region of the alphaD-helix reveals significant differences when compared with other TIR structures, especially the alphaD3-helix that corresponds to an insertion only present in plant TIR domains. Available mutagenesis data suggest that several conserved and exposed residues in this region are involved in the plant TIR signaling function.
Organizational Affiliation:
Infectious Diseases Center, Burnham Institute for Medical Research, La Jolla, California 92037, USA.