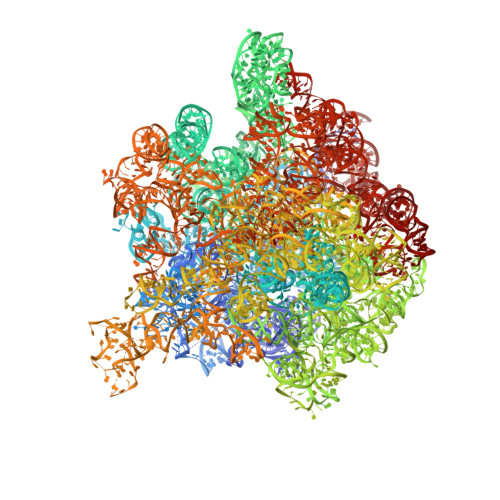
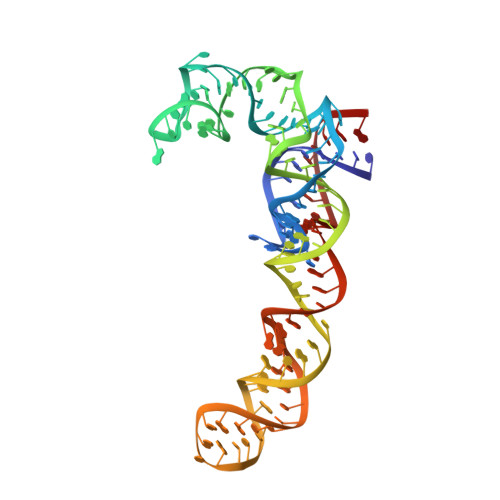
The structure of ribosome-lankacidin complex reveals ribosomal sites for synergistic antibiotics
Auerbach, T., Mermershtain, I., Davidovich, C., Bashan, A., Belousoff, M., Wekselman, I., Zimmerman, E., Xiong, L., Klepacki, D., Arakawa, K., Kinashi, H., Mankin, A.S., Yonath, A.(2010) Proc Natl Acad Sci U S A 107: 1983-1988
- PubMed: 20080686
- DOI: https://doi.org/10.1073/pnas.0914100107
- Primary Citation of Related Structures:
3JQ4 - PubMed Abstract:
Crystallographic analysis revealed that the 17-member polyketide antibiotic lankacidin produced by Streptomyces rochei binds at the peptidyl transferase center of the eubacterial large ribosomal subunit. Biochemical and functional studies verified this finding and showed interference with peptide bond formation. Chemical probing indicated that the macrolide lankamycin, a second antibiotic produced by the same species, binds at a neighboring site, at the ribosome exit tunnel. These two antibiotics can bind to the ribosome simultaneously and display synergy in inhibiting bacterial growth. The binding site of lankacidin and lankamycin partially overlap with the binding site of another pair of synergistic antibiotics, the streptogramins. Thus, at least two pairs of structurally dissimilar compounds have been selected in the course of evolution to act synergistically by targeting neighboring sites in the ribosome. These results underscore the importance of the corresponding ribosomal sites for development of clinically relevant synergistic antibiotics and demonstrate the utility of structural analysis for providing new directions for drug discovery.
Organizational Affiliation:
Department of Structural Biology, Weizmann Institute of Science, Rehovot 76100, Israel.