Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Ballandras-Colas, A., Brown, M., Cook, N.J., Dewdney, T.G., Demeler, B., Cherepanov, P., Lyumkis, D., Engelman, A.N.(2016) Nature 530: 358-361
- PubMed: 26887496
- DOI: https://doi.org/10.1038/nature16955
- Primary Citation of Related Structures:
3JCA, 5CZ1, 5CZ2, 5D7U - PubMed Abstract:
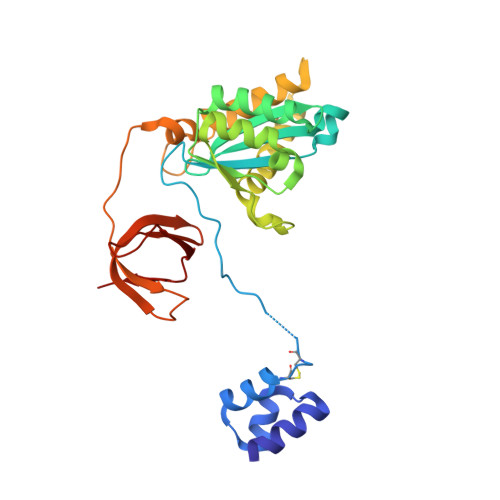
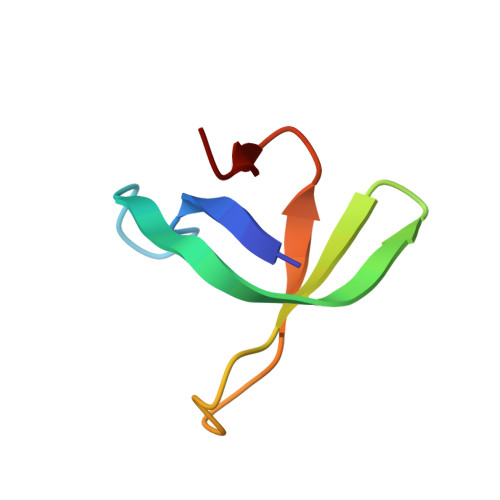
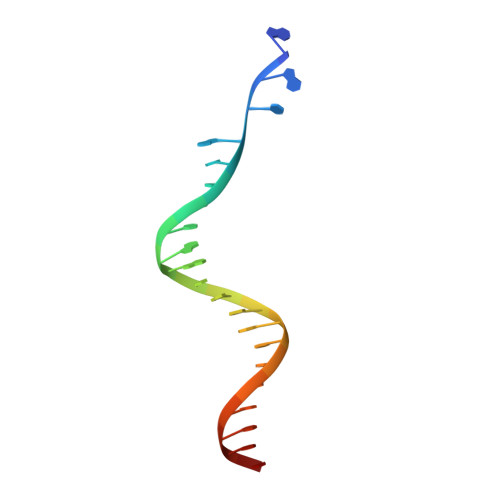
Retroviral integrase catalyses the integration of viral DNA into host target DNA, which is an essential step in the life cycle of all retroviruses. Previous structural characterization of integrase-viral DNA complexes, or intasomes, from the spumavirus prototype foamy virus revealed a functional integrase tetramer, and it is generally believed that intasomes derived from other retroviral genera use tetrameric integrase. However, the intasomes of orthoretroviruses, which include all known pathogenic species, have not been characterized structurally. Here, using single-particle cryo-electron microscopy and X-ray crystallography, we determine an unexpected octameric integrase architecture for the intasome of the betaretrovirus mouse mammary tumour virus. The structure is composed of two core integrase dimers, which interact with the viral DNA ends and structurally mimic the integrase tetramer of prototype foamy virus, and two flanking integrase dimers that engage the core structure via their integrase carboxy-terminal domains. Contrary to the belief that tetrameric integrase components are sufficient to catalyse integration, the flanking integrase dimers were necessary for mouse mammary tumour virus integrase activity. The integrase octamer solves a conundrum for betaretroviruses as well as alpharetroviruses by providing critical carboxy-terminal domains to the intasome core that cannot be provided in cis because of evolutionarily restrictive catalytic core domain-carboxy-terminal domain linker regions. The octameric architecture of the intasome of mouse mammary tumour virus provides new insight into the structural basis of retroviral DNA integration.
Organizational Affiliation:
Department of Cancer Immunology and Virology, Dana-Farber Cancer Institute and Department of Medicine, Harvard Medical School, 450 Brookline Avenue, Boston, Massachusetts 02215, USA.