Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization.
Sun, S., Xiang, Y., Akahata, W., Holdaway, H., Pal, P., Zhang, X., Diamond, M.S., Nabel, G.J., Rossmann, M.G.(2013) Elife 2: e00435-e00435
- PubMed: 23577234
- DOI: https://doi.org/10.7554/eLife.00435
- Primary Citation of Related Structures:
3J2W, 3J2X, 3J2Y, 3J2Z, 3J30, 4GQ9 - PubMed Abstract:
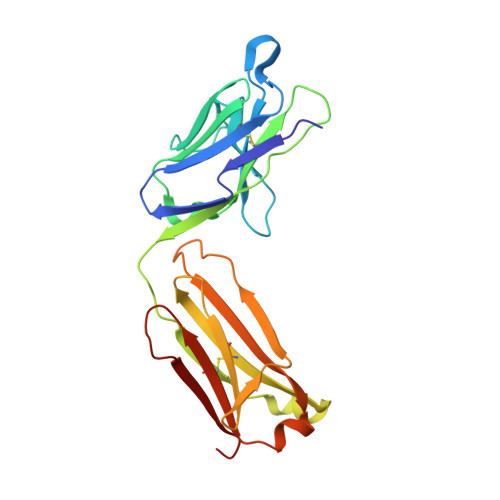
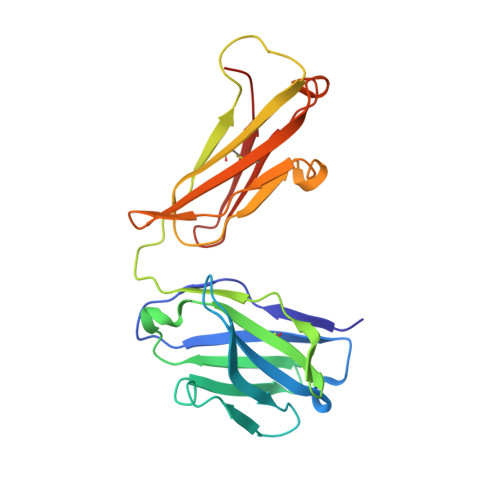
A 5.3 Å resolution, cryo-electron microscopy (cryoEM) map of Chikungunya virus-like particles (VLPs) has been interpreted using the previously published crystal structure of the Chikungunya E1-E2 glycoprotein heterodimer. The heterodimer structure was divided into domains to obtain a good fit to the cryoEM density. Differences in the T = 4 quasi-equivalent heterodimer components show their adaptation to different environments. The spikes on the icosahedral 3-fold axes and those in general positions are significantly different, possibly representing different phases during initial generation of fusogenic E1 trimers. CryoEM maps of neutralizing Fab fragments complexed with VLPs have been interpreted using the crystal structures of the Fab fragments and the VLP structure. Based on these analyses the CHK-152 antibody was shown to stabilize the viral surface, hindering the exposure of the fusion-loop, likely neutralizing infection by blocking fusion. The CHK-9, m10 and m242 antibodies surround the receptor-attachment site, probably inhibiting infection by blocking cell attachment. DOI:http://dx.doi.org/10.7554/eLife.00435.001.
Organizational Affiliation:
Department of Biological Sciences , Purdue University , West Lafayette , United States.