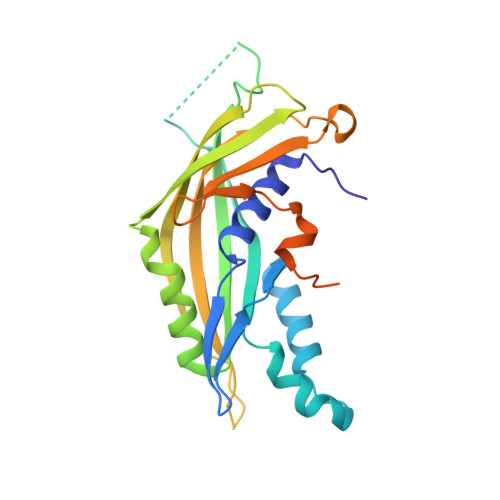
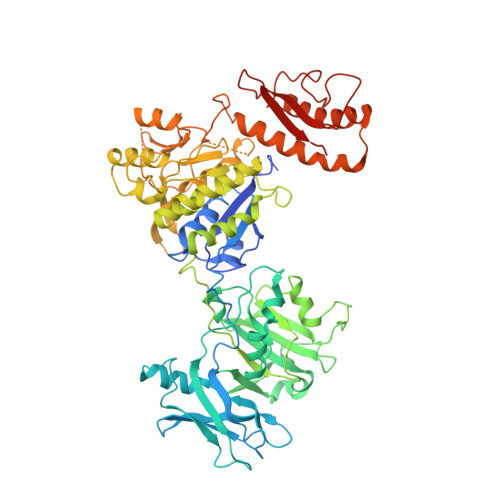
The molecular architecture of the bacteriophage t4 neck.
Fokine, A., Zhang, Z., Kanamaru, S., Bowman, V.D., Aksyuk, A.A., Arisaka, F., Rao, V.B., Rossmann, M.G.(2013) J Mol Biol 425: 1731-1744
- PubMed: 23434847
- DOI: https://doi.org/10.1016/j.jmb.2013.02.012
- Primary Citation of Related Structures:
3J2M, 3J2N, 3J2O, 4HUD, 4HUH - PubMed Abstract:
A hexamer of the bacteriophage T4 tail terminator protein, gp15, attaches to the top of the phage tail stabilizing the contractile sheath and forming the interface for binding of the independently assembled head. Here we report the crystal structure of the gp15 hexamer, describe its interactions in T4 virions that have either an extended tail or a contracted tail, and discuss its structural relationship to other phage proteins. The neck of T4 virions is decorated by the "collar" and "whiskers", made of fibritin molecules. Fibritin acts as a chaperone helping to attach the long tail fibers to the virus during the assembly process. The collar and whiskers are environment-sensing devices, regulating the retraction of the long tail fibers under unfavorable conditions, thus preventing infection. Cryo-electron microscopy analysis suggests that twelve fibritin molecules attach to the phage neck with six molecules forming the collar and six molecules forming the whiskers.
Organizational Affiliation:
Department of Biological Sciences, Purdue University, West Lafayette, IN 47907, USA.