tmRNA-SmpB: a journey to the centre of the bacterial ribosome.
Weis, F., Bron, P., Giudice, E., Rolland, J.P., Thomas, D., Felden, B., Gillet, R.(2010) EMBO J 29: 3810-3818
- PubMed: 20953161
- DOI: https://doi.org/10.1038/emboj.2010.252
- Primary Citation of Related Structures:
3IYQ, 3IYR - PubMed Abstract:
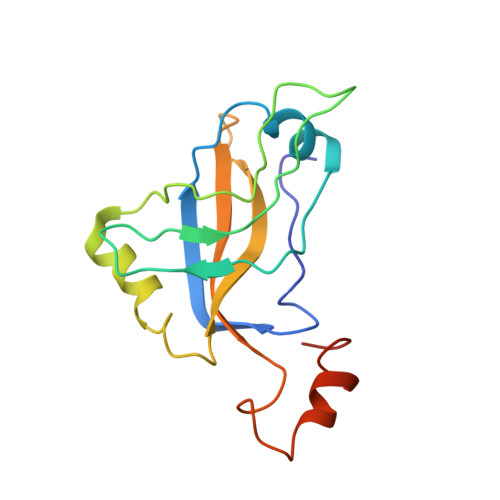
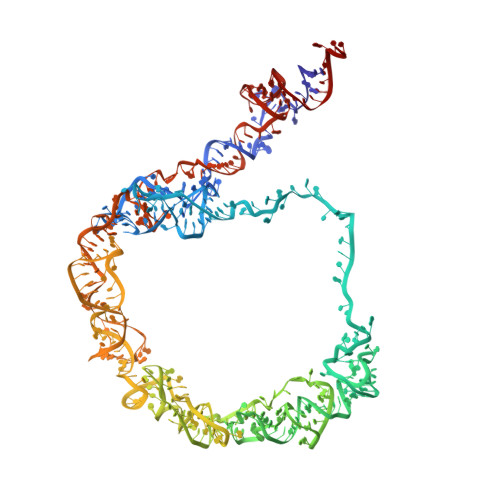
Ribosomes mediate protein synthesis by decoding the information carried by messenger RNAs (mRNAs) and catalysing peptide bond formation between amino acids. When bacterial ribosomes stall on incomplete messages, the trans-translation quality control mechanism is activated by the transfer-messenger RNA bound to small protein B (tmRNA-SmpB ribonucleoprotein complex). Trans-translation liberates the stalled ribosomes and triggers degradation of the incomplete proteins. Here, we present the cryo-electron microscopy structures of tmRNA-SmpB accommodated or translocated into stalled ribosomes. Two atomic models for each state are proposed. This study reveals how tmRNA-SmpB crosses the ribosome and how, as the problematic mRNA is ejected, the tmRNA resume codon is placed onto the ribosomal decoding site by new contacts between SmpB and the nucleotides upstream of the tag-encoding sequence. This provides a structural basis for the transit of the large tmRNA-SmpB complex through the ribosome and for the means by which the tmRNA internal frame is set for translation to resume.
Organizational Affiliation:
Université de Rennes, UMR CNRS Equipe Structure et Dynamique des Macromolécules, Rennes, France.