Adaptability of the ecdysone receptor bound to synthetic ligands
Moras, D., Billas, I.M.L., Browning, C.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Gene regulation protein | 263 | Heliothis virescens | Mutation(s): 0 Gene Names: ecr | 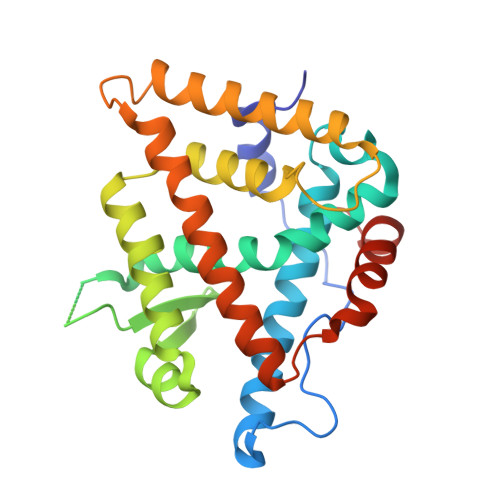 | |
UniProt | |||||
Find proteins for Q7SIF6 (Heliothis virescens) Explore Q7SIF6 Go to UniProtKB: Q7SIF6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q7SIF6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ecdysone receptor | B [auth D] | 265 | Helicoverpa armigera | Mutation(s): 4 Gene Names: usp | 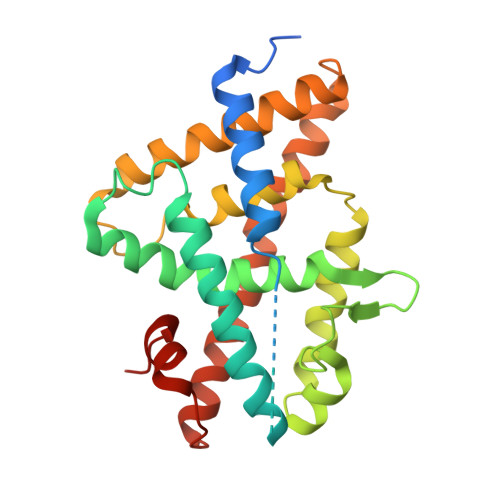 |
UniProt | |||||
Find proteins for B9UCQ4 (Helicoverpa armigera) Explore B9UCQ4 Go to UniProtKB: B9UCQ4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | B9UCQ4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| EPH Query on EPH | C [auth A] | L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE C39 H68 N O8 P MABRTXOVHMDVAT-AAEGOEIASA-N |  | ||
| 834 Query on 834 | D | N-[2-(2-chlorophenyl)-4-methyl-5-(1-methylethyl)-1H-imidazol-1-yl]-5-methyl-2,3-dihydro-1,4-benzodioxine-6-carboxamide C23 H24 Cl N3 O3 XLNQTHXOKGRTAL-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 147.875 | α = 90 |
| b = 147.875 | β = 90 |
| c = 59.776 | γ = 120 |
| Software Name | Purpose |
|---|---|
| CNS | refinement |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data collection |
| DENZO | data reduction |
| SCALEPACK | data scaling |
| HKL-2000 | data scaling |
| AMoRE | phasing |