Crystal structure of Two component response regulator from Cytophaga hutchinsonii
Syed Ibrahim, B., Burley, S.K., Swaminathan, S.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Two component response regulator | 146 | Cytophaga hutchinsonii ATCC 33406 | Mutation(s): 0 Gene Names: narL, CHU_1356 | 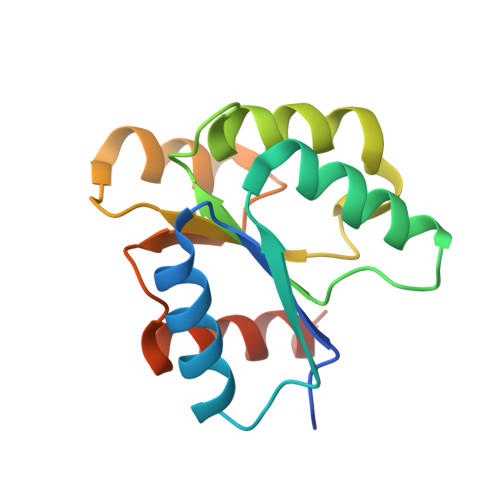 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 63.671 | α = 90 |
| b = 63.671 | β = 90 |
| c = 158.388 | γ = 120 |
| Software Name | Purpose |
|---|---|
| CBASS | data collection |
| SHELX | model building |
| SHARP | phasing |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| SCALEPACK | data scaling |
| SHELX | phasing |