X-ray crystallographic structure of Inorganic Pyrophosphatase at 298K from archaeon Thermococcus thioreducens
Hughes, R.C., Coates, L., Meehan, E.J., Ng, J.D.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Th-IPP | 178 | Thermococcus thioreducens | Mutation(s): 0 | 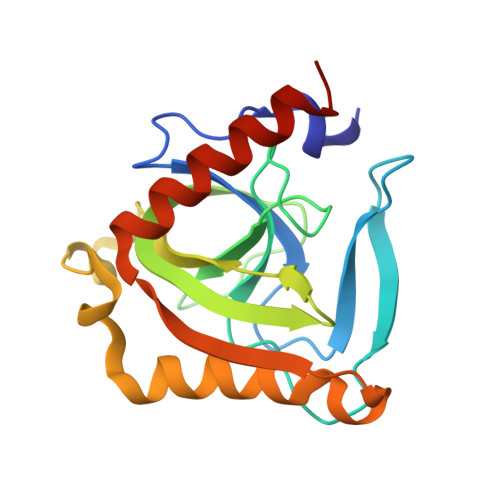 | |
UniProt | |||||
Find proteins for H0USY5 (Thermococcus thioreducens) Explore H0USY5 Go to UniProtKB: H0USY5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | H0USY5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MPD Query on MPD | J [auth C] | (4S)-2-METHYL-2,4-PENTANEDIOL C6 H14 O2 SVTBMSDMJJWYQN-YFKPBYRVSA-N |  | ||
| MRD Query on MRD | G [auth A], P [auth F] | (4R)-2-METHYLPENTANE-2,4-DIOL C6 H14 O2 SVTBMSDMJJWYQN-RXMQYKEDSA-N |  | ||
| ACE Query on ACE | M [auth E] | ACETYL GROUP C2 H4 O IKHGUXGNUITLKF-UHFFFAOYSA-N |  | ||
| CA Query on CA | H [auth A] I [auth B] K [auth C] L [auth D] N [auth E] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 106.112 | α = 90 |
| b = 95.464 | β = 98.12 |
| c = 113.677 | γ = 90 |
| Software Name | Purpose |
|---|---|
| DENZO | data reduction |
| SCALEPACK | data scaling |
| MOLREP | phasing |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| SERGUI | data collection |