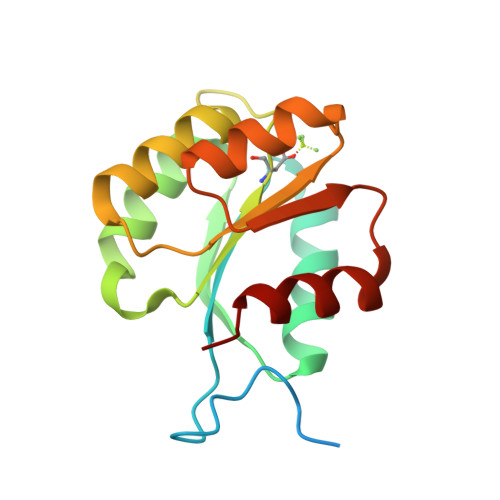
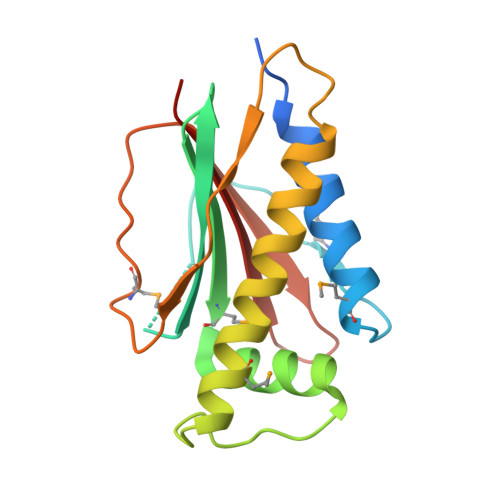
Identical phosphatase mechanisms achieved through distinct modes of binding phosphoprotein substrate.
Pazy, Y., Motaleb, M.A., Guarnieri, M.T., Charon, N.W., Zhao, R., Silversmith, R.E.(2010) Proc Natl Acad Sci U S A 107: 1924-1929
- PubMed: 20080618
- DOI: https://doi.org/10.1073/pnas.0911185107
- Primary Citation of Related Structures:
3HZH - PubMed Abstract:
Two-component signal transduction systems are widespread in prokaryotes and control numerous cellular processes. Extensive investigation of sensor kinase and response regulator proteins from many two-component systems has established conserved sequence, structural, and mechanistic features within each family. In contrast, the phosphatases which catalyze hydrolysis of the response regulator phosphoryl group to terminate signal transduction are poorly understood. Here we present structural and functional characterization of a representative of the CheC/CheX/FliY phosphatase family. The X-ray crystal structure of Borrelia burgdorferi CheX complexed with its CheY3 substrate and the phosphoryl analogue reveals a binding orientation between a response regulator and an auxiliary protein different from that shared by every previously characterized example. The surface of CheY3 containing the phosphoryl group interacts directly with a long helix of CheX which bears the conserved (E - X(2) - N) motif. Conserved CheX residues Glu96 and Asn99, separated by a single helical turn, insert into the CheY3 active site. Structural and functional data indicate that CheX Asn99 and CheY3 Thr81 orient a water molecule for hydrolytic attack. The catalytic residues of the CheX.CheY3 complex are virtually superimposable on those of the Escherichia coli CheZ phosphatase complexed with CheY, even though the active site helices of CheX and CheZ are oriented nearly perpendicular to one other. Thus, evolution has found two structural solutions to achieve the same catalytic mechanism through different helical spacing and side chain lengths of the conserved acid/amide residues in CheX and CheZ.
Organizational Affiliation:
Department of Microbiology and Immunology, University of North Carolina, Chapel Hill, NC 27599, USA.