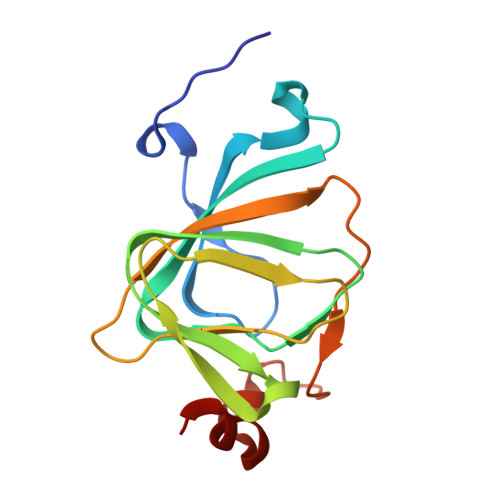
The polyketide cyclase RemF from Streptomyces resistomycificus contains an unusual octahedral zinc binding site
Silvennoinen, L., Sandalova, T., Schneider, G.(2009) FEBS Lett 583: 2917-2921
- PubMed: 19665022
- DOI: https://doi.org/10.1016/j.febslet.2009.07.061
- Primary Citation of Related Structures:
3HT1, 3HT2 - PubMed Abstract:
RemF is a polyketide cyclase involved in the biosynthesis of the aromatic pentacyclic metabolite resistomycin in Streptomyces resistomycificus. The enzyme is a member of a structurally hitherto uncharacterized class of polyketide cyclases. The crystal structure of RemF was determined by SAD and refined to 1.2 A resolution. The enzyme subunit shows a beta-sandwich structure with a topology characteristic for the cupin fold. RemF contains a metal binding site located at the bottom of the predominantly hydrophobic active site cavity. A zinc ion is coordinated to four histidine side chains, and two water molecules in octahedral ligand sphere geometry, highly unusual for zinc binding sites in proteins.
Organizational Affiliation:
Department of Medical Biophysics and Biochemistry, Karolinska Institutet, Stockholm, Sweden.