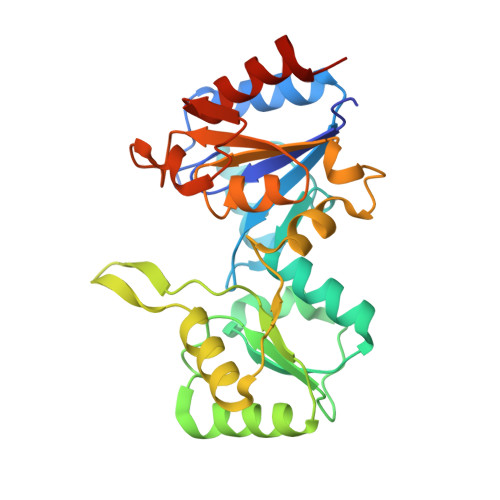
The crystal structure of human haloacid dehalogenase-like hydrolase domain containing 2 (HDHD2)
Ugochukwu, E., Shafqat, N., Yue, W.W., Cocking, R., Bray, J.E., Muniz, J.R.C., Krojer, T., von Delft, F., Bountra, C., Arrowsmith, C.H., Weigelt, J., Edwards, A., Oppermann, U., Structural Genomics Consortium (SGC)To be published.