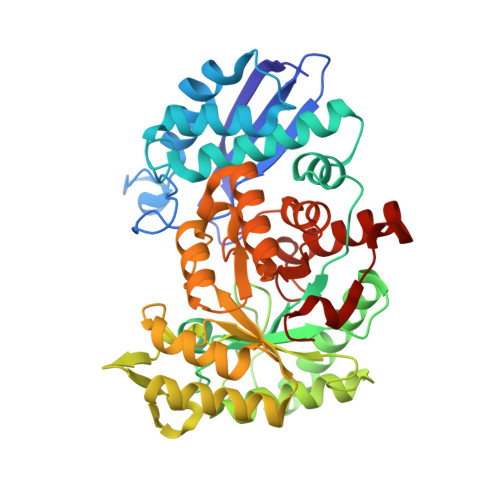
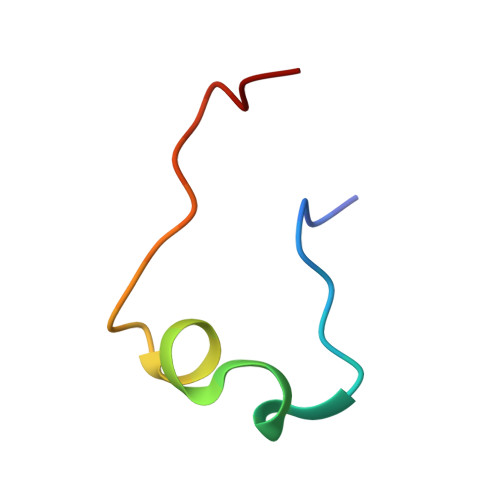
Molecular recognition between Escherichia coli enolase and ribonuclease E.
Nurmohamed, S., McKay, A.R., Robinson, C.V., Luisi, B.F.(2010) Acta Crystallogr D Biol Crystallogr 66: 1036-1040
- PubMed: 20823555
- DOI: https://doi.org/10.1107/S0907444910030015
- Primary Citation of Related Structures:
3H8A - PubMed Abstract:
In Escherichia coli and many other bacterial species, the glycolytic enzyme enolase is a component of the multi-enzyme RNA degradosome, an assembly that is involved in RNA processing and degradation. Enolase is recruited into the degradosome through interactions with a small recognition motif located within the degradosome-scaffolding domain of RNase E. Here, the crystal structure of enolase bound to its cognate site from RNase E (residues 823-850) at 1.9 A resolution is presented. The structure suggests that enolase may help to organize an adjacent conserved RNA-binding motif in RNase E.
Organizational Affiliation:
Department of Biochemistry, University of Cambridge, England.