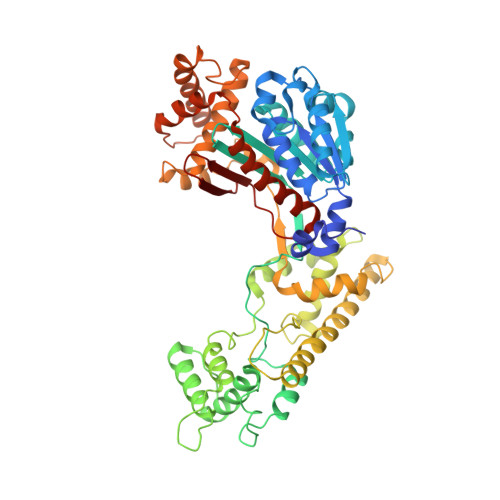
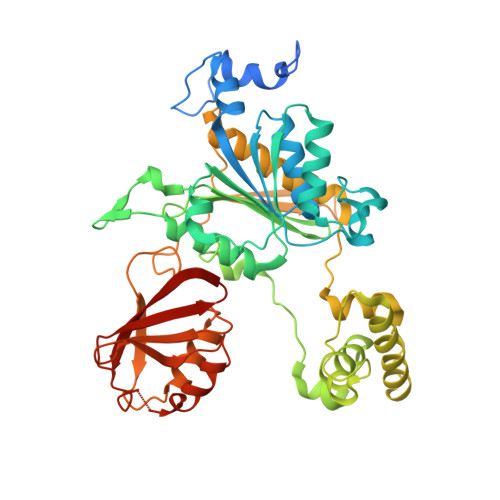
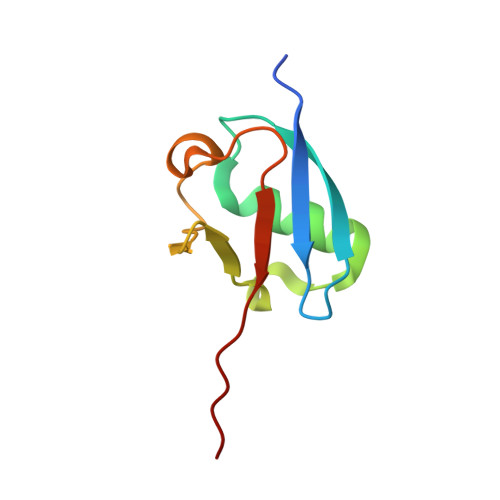
Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ.
Brownell, J.E., Sintchak, M.D., Gavin, J.M., Liao, H., Bruzzese, F.J., Bump, N.J., Soucy, T.A., Milhollen, M.A., Yang, X., Burkhardt, A.L., Ma, J., Loke, H.K., Lingaraj, T., Wu, D., Hamman, K.B., Spelman, J.J., Cullis, C.A., Langston, S.P., Vyskocil, S., Sells, T.B., Mallender, W.D., Visiers, I., Li, P., Claiborne, C.F., Rolfe, M., Bolen, J.B., Dick, L.R.(2010) Mol Cell 37: 102-111
- PubMed: 20129059
- DOI: https://doi.org/10.1016/j.molcel.2009.12.024
- Primary Citation of Related Structures:
3GZN - PubMed Abstract:
The NEDD8-activating enzyme (NAE) initiates a protein homeostatic pathway essential for cancer cell growth and survival. MLN4924 is a selective inhibitor of NAE currently in clinical trials for the treatment of cancer. Here, we show that MLN4924 is a mechanism-based inhibitor of NAE and creates a covalent NEDD8-MLN4924 adduct catalyzed by the enzyme. The NEDD8-MLN4924 adduct resembles NEDD8 adenylate, the first intermediate in the NAE reaction cycle, but cannot be further utilized in subsequent intraenzyme reactions. The stability of the NEDD8-MLN4924 adduct within the NAE active site blocks enzyme activity, thereby accounting for the potent inhibition of the NEDD8 pathway by MLN4924. Importantly, we have determined that compounds resembling MLN4924 demonstrate the ability to form analogous adducts with other ubiquitin-like proteins (UBLs) catalyzed by their cognate-activating enzymes. These findings reveal insights into the mechanism of E1s and suggest a general strategy for selective inhibition of UBL conjugation pathways.
Organizational Affiliation:
Discovery, Millennium Pharmaceuticals, Inc., 40 Landsdowne Street, Cambridge, MA 02139, USA.