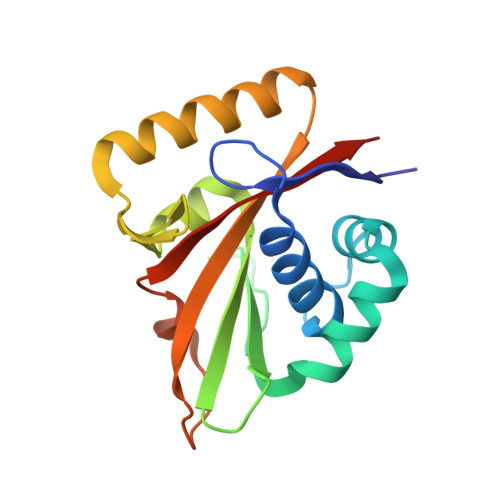
The structure of Mg-ATPase nucleotide-binding domain at 1.6 A resolution reveals a unique ATP-binding motif
Hakansson, K.O.(2009) Acta Crystallogr D Biol Crystallogr 65: 1181-1186
- PubMed: 19923713
- DOI: https://doi.org/10.1107/S090744490903306X
- Primary Citation of Related Structures:
3GWI - PubMed Abstract:
The structure of the nucleotide-binding domain of the Mg-ATPase MgtA from Escherichia coli has been solved and refined to 1.6 A resolution. The structure is made up of a six-stranded beta-sheet and a bundle of three alpha-helices, with the nucleotide-binding site sandwiched in between. The MgtA nucleotide-binding domain is shorter and more compact compared with that of the related Ca-ATPase and lacks one of the beta-strands at the edge of the beta-sheet. The ATP-binding pocket is surrounded by three sequence and structural motifs known from other P-type ATPases and a fourth unique motif that is found only in Mg-ATPases. This motif consists of a short polypeptide stretch running very close to the ATP-binding site, while in Ca-ATPase the binding site is more open, with the corresponding polypeptide segment folded away from the active site.
Organizational Affiliation:
Department of Biology, August Krogh Building, University of Copenhagen, DK-2100 Copenhagen O, Denmark. kohakansson@bio.ku.dk