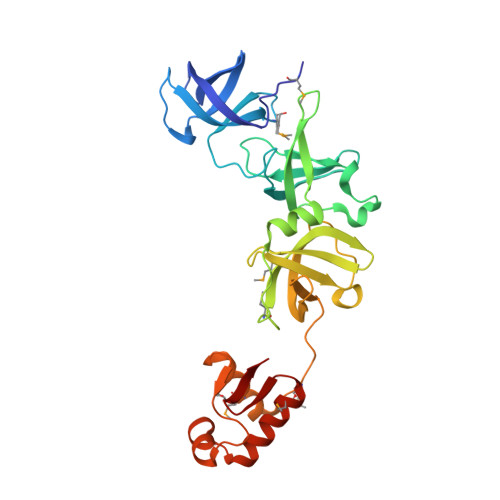
Structure of a virulence regulatory factor CvfB reveals a novel winged helix RNA binding module.
Matsumoto, Y., Xu, Q., Miyazaki, S., Kaito, C., Farr, C.L., Axelrod, H.L., Chiu, H.J., Klock, H.E., Knuth, M.W., Miller, M.D., Elsliger, M.A., Deacon, A.M., Godzik, A., Lesley, S.A., Sekimizu, K., Wilson, I.A.(2010) Structure 18: 537-547
- PubMed: 20399190
- DOI: https://doi.org/10.1016/j.str.2010.02.007
- Primary Citation of Related Structures:
3GO5 - PubMed Abstract:
CvfB is a conserved regulatory protein important for the virulence of Staphylococcus aureus. We show here that CvfB binds RNA. The crystal structure of the CvfB ortholog from Streptococcus pneumoniae at 1.4 A resolution reveals a unique RNA binding protein that is formed from a concatenation of well-known structural modules that bind nucleic acids: three consecutive S1 RNA binding domains and a winged helix (WH) domain. The third S1 and the WH domains are required for cooperative RNA binding and form a continuous surface that likely contributes to the RNA interaction. The WH domain is critical to CvfB function and contains a unique sequence motif. Thus CvfB represents a novel assembly of modules for binding RNA.
Organizational Affiliation:
Laboratory of Microbiology, Graduate School of Pharmaceutical Sciences, The University of Tokyo, Tokyo 113-0033, Japan.