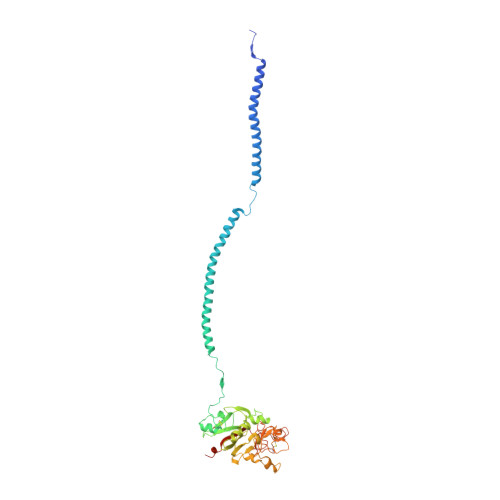
Crystal structure of human fibrinogen.
Kollman, J.M., Pandi, L., Sawaya, M.R., Riley, M., Doolittle, R.F.(2009) Biochemistry 48: 3877-3886
- PubMed: 19296670
- DOI: https://doi.org/10.1021/bi802205g
- Primary Citation of Related Structures:
3GHG - PubMed Abstract:
A crystal structure of human fibrinogen has been determined at approximately 3.3 A resolution. The protein was purified from human blood plasma, first by a cold ethanol precipitation procedure and then by stepwise chromatography on DEAE-cellulose. A product was obtained that was homogeneous on SDS-polyacrylamide gels. Nonetheless, when individual crystals used for X-ray diffraction were examined by SDS gel electrophoresis after data collection, two species of alpha chain were present, indicating that some proteolysis had occurred during the course of operations. Amino-terminal sequencing on post-X-ray crystals showed mostly intact native alpha- and gamma-chain sequences (the native beta chain is blocked). The overall structure differs from that of a native fibrinogen from chicken blood and those reported for a partially proteolyzed bovine fibrinogen in the nature of twist in the coiled-coil regions, likely due to weak forces imparted by unique crystal packing. As such, the structure adds to the inventory of possible conformations that may occur in solution. Other features include a novel interface with an antiparallel arrangement of beta chains and a unique tangential association of coiled coils from neighboring molecules. The carbohydrate groups attached to beta chains are unusually prominent, the full sweep of 11 sugar residues being positioned. As was the case for native chicken fibrinogen, no resolvable electron density could be associated with alphaC domains.
Organizational Affiliation:
Department of Chemistry and Biochemistry and Division of Biology, University of California at San Diego, La Jolla, California 92093-0314, USA.