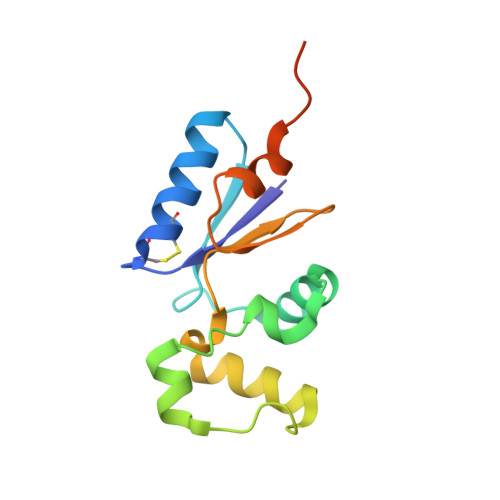
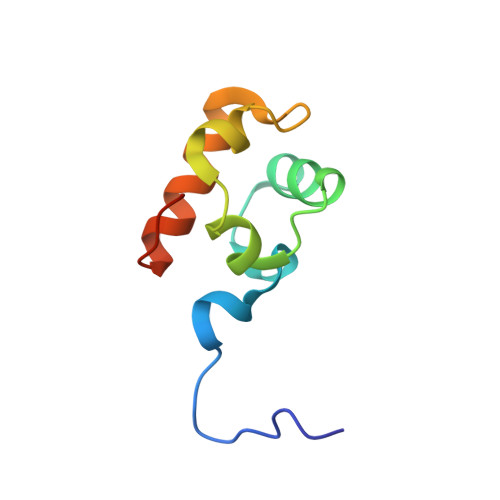
Crystal structure of the in vivo-assembled Bacillus subtilis Spx/RNA polymerase alpha subunit C-terminal domain complex
Lamour, V., Westblade, L.F., Campbell, E.A., Darst, S.A.(2009) J Struct Biol 168: 352-356
- PubMed: 19580872
- DOI: https://doi.org/10.1016/j.jsb.2009.07.001
- Primary Citation of Related Structures:
3GFK - PubMed Abstract:
The Bacillus subtilis Spx protein is a global transcription factor that interacts with the C-terminal domain of the RNA polymerase alpha subunit (alphaCTD) and regulates transcription of genes involved in thiol-oxidative stress, sporulation, competence, and organosulfur metabolism. Here we determined the X-ray crystal structure of the Spx/alphaCTD complex from an entirely new crystal form than previously reported [Newberry, K.J., Nakano, S., Zuber, P., Brennan, R.G., 2005. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc. Natl. Acad. Sci. USA 102, 15839-15844]. Comparison of the previously reported sulfate-bound complex and our sulfate-free complex reveals subtle conformational changes that may be important for the role of Spx in regulating organosulfur metabolism.
Organizational Affiliation:
The Rockefeller University, New York, NY 10065, USA.