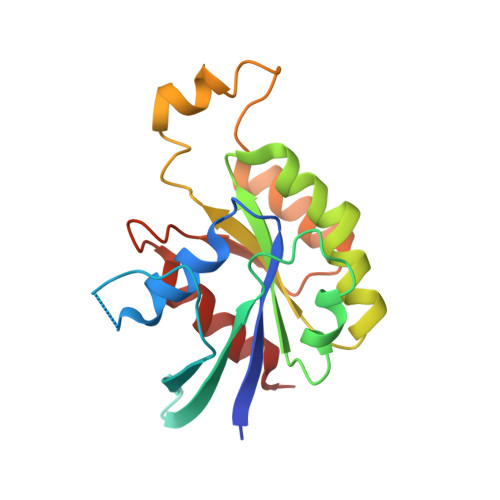
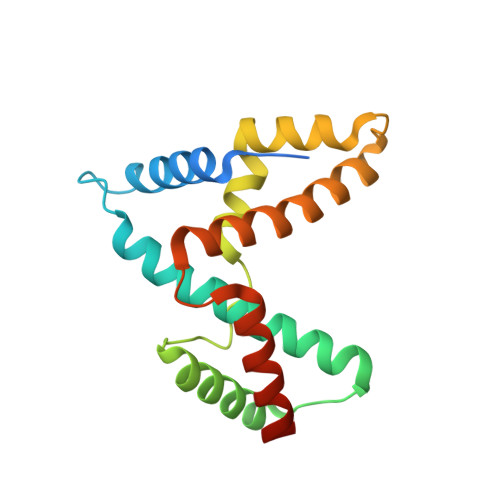
Structural insights into host GTPase isoform selection by a family of bacterial GEF mimics
Huang, Z., Sutton, S.E., Wallenfang, A.J., Orchard, R.C., Wu, X., Feng, Y., Chai, J., Alto, N.M.(2009) Nat Struct Mol Biol 16: 853-860
- PubMed: 19620963
- DOI: https://doi.org/10.1038/nsmb.1647
- Primary Citation of Related Structures:
3GCG - PubMed Abstract:
The Escherichia coli type III effector Map belongs to a large family of bacterial virulence factors that activate host Rho GTPase signaling pathways through an unknown molecular mechanism. Here we report direct evidence that Map functions as a potent and selective guanine-nucleotide exchange factor (GEF) for Cdc42. The 2.3-A structure of the Map-Cdc42 complex revealed that Map mimics the GEF strategy of the mammalian Dbl family but has a three-dimensional architecture that is nearly identical to the bacterial GEF Salmonella spp. SopE. A comparative analysis between human and bacterial GEFs revealed a previously uncharacterized pairing mechanism between Map and the variable beta2-3 interswitch region of Cdc42. We propose a GTPase selection model that is experimentally validated by the preferential activation Rac1 and RhoA by the Shigella spp. effectors IpgB1 and IpgB2, respectively. These results significantly expand the repertoire of bacterial GEF mimics and unify a GEF selection mechanism for host GTPase substrates.
Organizational Affiliation:
National Institute of Biological Sciences, Zhong Guan Cun Life Science Park, Beijing, China.