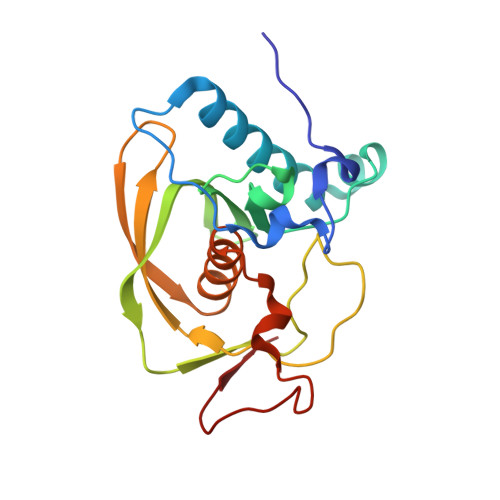
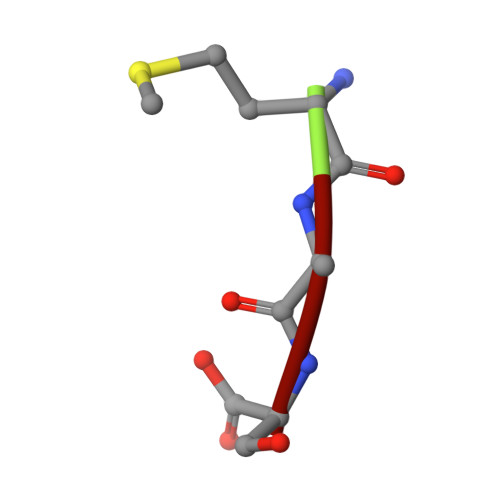
Crystal structure of an EfPDF complex with Met-Ala-Ser based on crystallographic packing.
Nam, K.H., Kim, K.H., Kim, E.E., Hwang, K.Y.(2009) Biochem Biophys Res Commun 381: 630-633
- PubMed: 19249287
- DOI: https://doi.org/10.1016/j.bbrc.2009.02.113
- Primary Citation of Related Structures:
3G6N - PubMed Abstract:
PDF (peptide deformylase) plays a critical role in the production of mature proteins by removing the N-formyl polypeptide of nascent proteins in the prokaryote cell system. This protein is essential for bacterial growth, making it an attractive target for the design of new antibiotics. Accordingly, PDF has been evaluated as a drug target; however, architectural mechanism studies of PDF have not yet fully elucidated its molecular function. We recently reported the crystal structure of PDF produced by Enterococcus faecium [K.H. Nam, J.I. Ham, A. Priyadarshi, E.E. Kim, N. Chung, K.Y. Hwang, "Insight into the antibacterial drug design and architectural mechanism of peptide recognition from the E. faecium peptide deformylase structure", Proteins 74 (2009) 261-265]. Here, we present the crystal structure of the EfPDF complex with MAS (Met-Ser-Ala), thereby not only delineating the architectural mechanism for the recognition of mimic-peptides by N-terminal cleaved expression peptide, but also suggesting possible targets for rational design of antibacterial drugs. In addition to their implications for drug design, these structural studies will facilitate elucidation of the architectural mechanism responsible for the peptide recognition of PDF.
Organizational Affiliation:
Korea University, Seoul, South Korea.