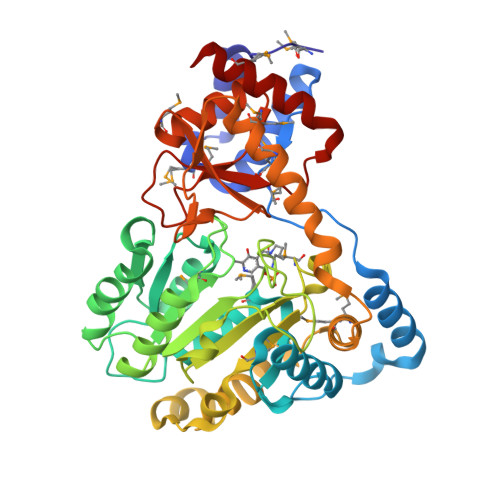
Molecular characterization of novel pyridoxal-5'-phosphate-dependent enzymes from the human microbiome.
Fleischman, N.M., Das, D., Kumar, A., Xu, Q., Chiu, H.J., Jaroszewski, L., Knuth, M.W., Klock, H.E., Miller, M.D., Elsliger, M.A., Godzik, A., Lesley, S.A., Deacon, A.M., Wilson, I.A., Toney, M.D.(2014) Protein Sci 23: 1060-1076
- PubMed: 24888348
- DOI: https://doi.org/10.1002/pro.2493
- Primary Citation of Related Structures:
3G0T - PubMed Abstract:
Pyridoxal-5'-phosphate or PLP, the active form of vitamin B6, is a highly versatile cofactor that participates in a large number of mechanistically diverse enzymatic reactions in basic metabolism. PLP-dependent enzymes account for ∼1.5% of most prokaryotic genomes and are estimated to be involved in ∼4% of all catalytic reactions, making this an important class of enzymes. Here, we structurally and functionally characterize three novel PLP-dependent enzymes from bacteria in the human microbiome: two are from Eubacterium rectale, a dominant, nonpathogenic, fecal, Gram-positive bacteria, and the third is from Porphyromonas gingivalis, which plays a major role in human periodontal disease. All adopt the Type I PLP-dependent enzyme fold and structure-guided biochemical analysis enabled functional assignments as tryptophan, aromatic, and probable phosphoserine aminotransferases.
Organizational Affiliation:
Department of Chemistry, University of California, Davis, California, 95616.