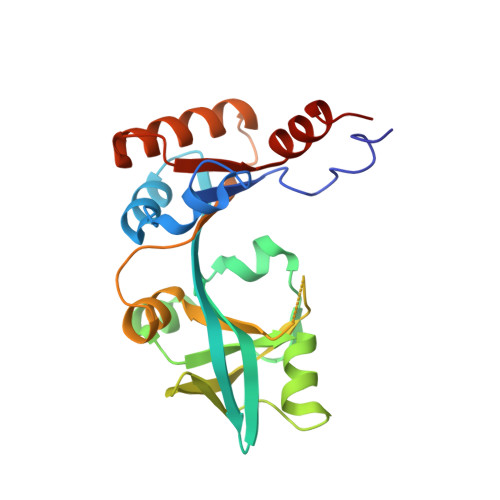
Crystal structure of the periplasmic region of MacB, a noncanonic ABC transporter
Xu, Y., Sim, S.-H., Nam, K.H., Jin, X.L., Kim, H.-M., Hwang, K.Y., Lee, K., Ha, N.-C.(2009) Biochemistry 48: 5218-5225
- PubMed: 19432486
- DOI: https://doi.org/10.1021/bi900415t
- Primary Citation of Related Structures:
3FTJ - PubMed Abstract:
MacB is a noncanonic ABC-type transporter within Gram-negative bacteria, which is responsible both for the efflux of macrolide antibiotics and for the secretion of heat-stable enterotoxin II. In Escherichia coli, MacB requires the membrane fusion protein MacA and the multifunctional outer membrane channel TolC to pump substrates to the external medium. Sequence analysis of MacB suggested that MacB has a relatively large periplasmic region. To gain insight into how MacB assembles with MacA and TolC, we determined the crystal structure of the periplasmic region of Actinobacillus actinomycetemcomitans MacB. Fold matching program reveals that parts of the MacB periplasmic region have structural motifs in common with the RND-type transporter AcrB. Since it behaved as a monomer in solution, our finding is consistent with the dimeric nature of full-length MacB, providing an insight into the assembly in the tripartite efflux pump.
Organizational Affiliation:
College of Pharmacy and Research Institute for Drug Development, Pusan National University, Busan 609-735, Republic of Korea.