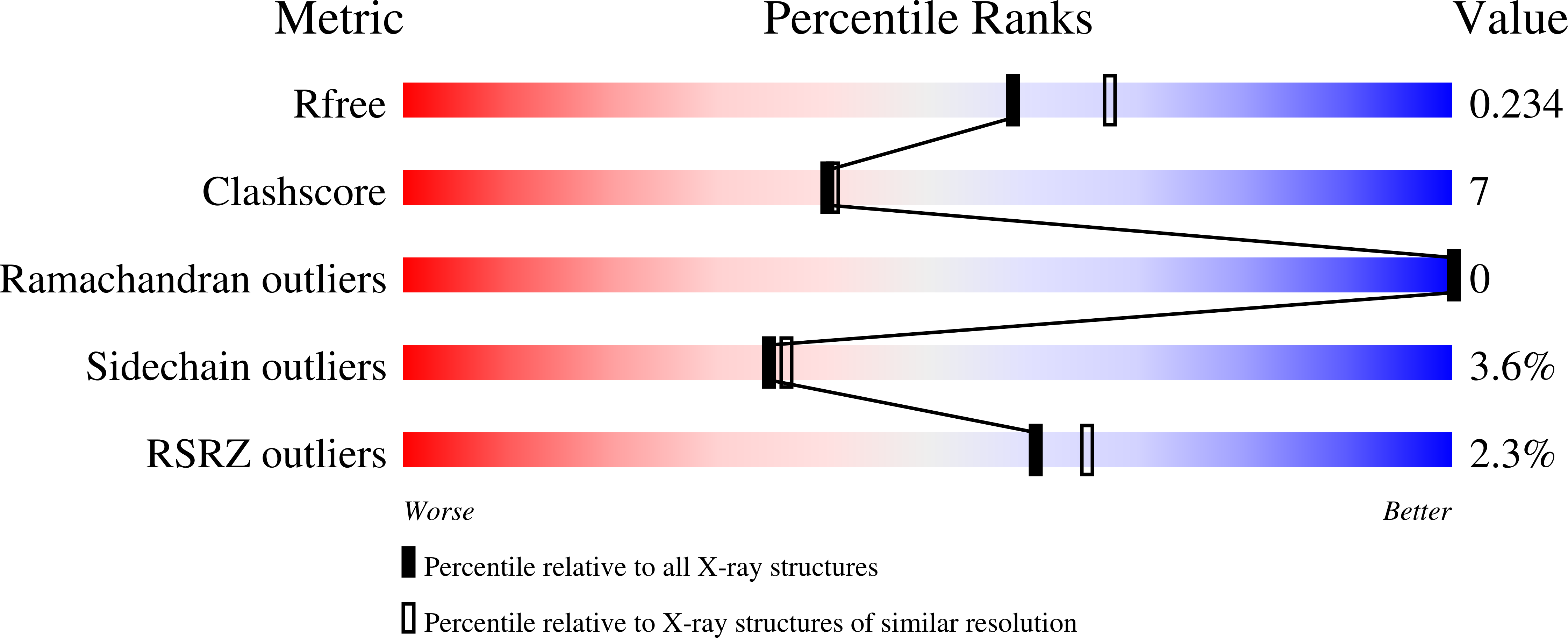
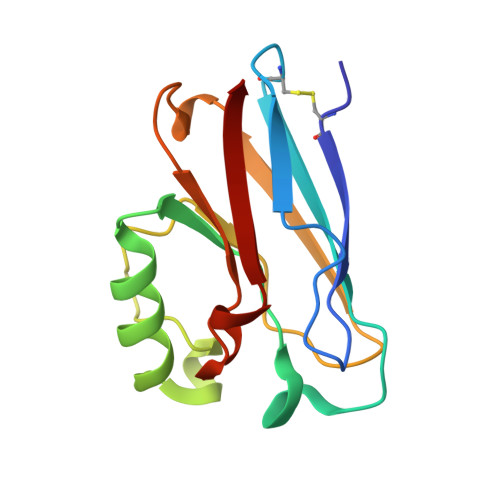
Type Zero Copper Proteins.
Lancaster, K.M., Debeer George, S., Yokoyama, K., Richards, J.H., Gray, H.B.(2009) Nat Chem 1: 711-715
- PubMed: 20305734
- DOI: https://doi.org/10.1038/nchem.412
- Primary Citation of Related Structures:
3FPY, 3FQ1, 3FQ2, 3FQY - PubMed Abstract:
Many proteins contain copper in a range of coordination environments, where it has various biological roles, such as transferring electrons or activating dioxygen. These copper sites can be classified by their function or spectroscopic properties. Those with a single copper atom are either type 1, with an intense absorption band near 600 nm, or type 2, with weak absorption in the visible region. We have built a novel copper(II) binding site within structurally modified Pseudomonas aeruginosa azurins that does not resemble either existing type, which we therefore call 'type zero'. X-ray crystallographic analysis shows that these sites adopt distorted tetrahedral geometries, with an unusually short Cu–O (G45 carbonyl) bond. Relatively weak absorption near 800 nm and narrow parallel hyperfine splittings in electron paramagnetic resonance spectra are the spectroscopic signatures of type zero copper. Cyclic voltammetric experiments demonstrate that the electron transfer reactivities of type-zero azurins are enhanced relative to that of the corresponding type 2 (C112D) protein.
Organizational Affiliation:
California Institute of Technology, Pasadena, California 91125, USA.