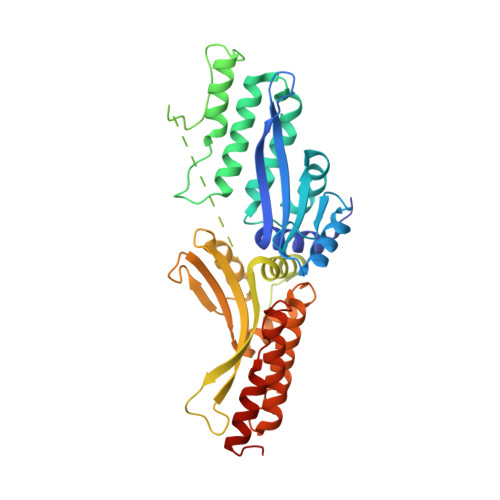
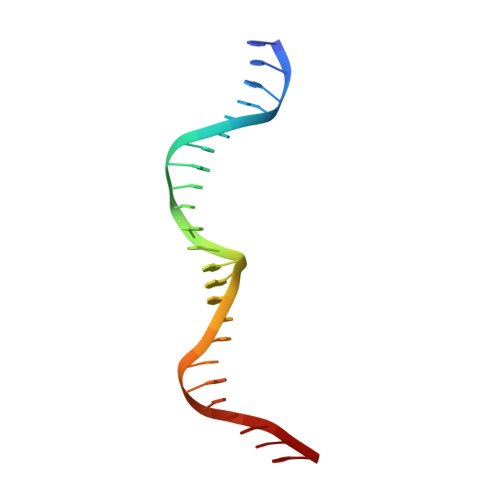
Generation of single-chain LAGLIDADG homing endonucleases from native homodimeric precursor proteins.
Li, H., Pellenz, S., Ulge, U., Stoddard, B.L., Monnat, R.J.(2009) Nucleic Acids Res 37: 1650-1662
- PubMed: 19153140
- DOI: https://doi.org/10.1093/nar/gkp004
- Primary Citation of Related Structures:
3FD2 - PubMed Abstract:
Homing endonucleases (HEs) cut long DNA target sites with high specificity to initiate and target the lateral transfer of mobile introns or inteins. This high site specificity of HEs makes them attractive reagents for gene targeting to promote DNA modification or repair. We have generated several hundred catalytically active, monomerized versions of the well-characterized homodimeric I-CreI and I-MsoI LAGLIDADG family homing endonuclease (LHE) proteins. Representative monomerized I-CreI and I-MsoI proteins (collectively termed mCreIs or mMsoIs) were characterized in detail by using a combination of biochemical, biophysical and structural approaches. We also demonstrated that both mCreI and mMsoI proteins can promote cleavage-dependent recombination in human cells. The use of single chain LHEs should simplify gene modification and targeting by requiring the expression of a single small protein in cells, rather than the coordinate expression of two separate protein coding genes as is required when using engineered heterodimeric zinc finger or homing endonuclease proteins.
Organizational Affiliation:
Department of Pathology, University of Washington, Box 357705, Seattle, WA 98195, USA.