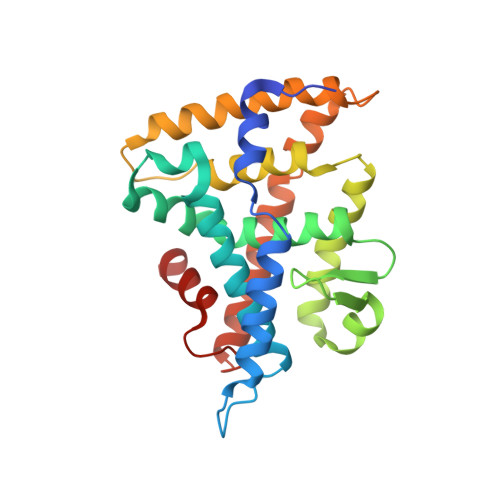
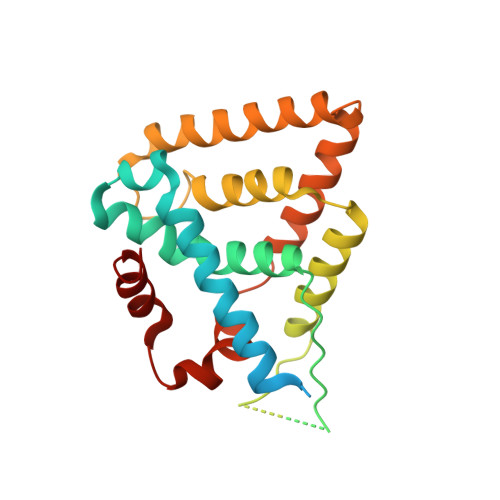
The structure of corepressor Dax-1 bound to its target nuclear receptor LRH-1.
Sablin, E.P., Woods, A., Krylova, I.N., Hwang, P., Ingraham, H.A., Fletterick, R.J.(2008) Proc Natl Acad Sci U S A 105: 18390-18395
- PubMed: 19015525
- DOI: https://doi.org/10.1073/pnas.0808936105
- Primary Citation of Related Structures:
3F5C - PubMed Abstract:
The Dax-1 protein is an enigmatic nuclear receptor that lacks an expected DNA binding domain, yet functions as a potent corepressor of nuclear receptors. Here we report the structure of Dax-1 bound to one of its targets, liver receptor homolog 1 (LRH-1). Unexpectedly, Dax-1 binds to LRH-1 using a new module, a repressor helix built from a family conserved sequence motif, PCFXXLP. Mutations in this repressor helix that are linked with human endocrine disorders dissociate the complex and attenuate Dax-1 function. The structure of the Dax-1:LRH-1 complex provides the molecular mechanism for the function of Dax-1 as a potent transcriptional repressor.
Organizational Affiliation:
Department of Biochemistry and Biophysics, University of California, San Francisco, CA 94143, USA.