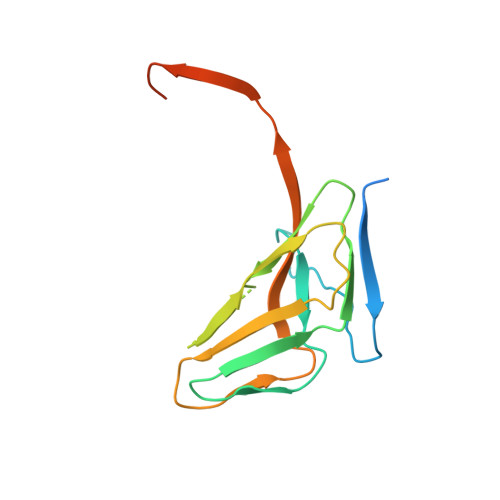
Molecular shape and prominent role of beta-strand swapping in organization of dUTPase oligomers.
Takacs, E., Barabas, O., Petoukhov, M.V., Svergun, D.I., Vertessy, B.G.(2009) FEBS Lett 583: 865-871
- PubMed: 19302784
- DOI: https://doi.org/10.1016/j.febslet.2009.02.011
- Primary Citation of Related Structures:
3ECY - PubMed Abstract:
Most dUTP pyrophosphatases (dUTPases) are homotrimers with interfaces formed between subunit surfaces, in the central channel, and by C-terminal beta-strand swapping. Analysis of intersubunit interactions reveals an important cohesive role for the C-terminus. This is reflected in the crystal structure of fruitfly dUTPase displaying a dimeric organization in crystals grown in alcohol solution, where only beta-strand swapping interactions between subunits are retained from the usual trimer structure. Mutations of a suggested hinge proline destabilize human and Escherichia coli dUTPases without preventing trimeric organization. Trimer formation was, however, prevented in the human enzyme by truncating the C-terminus before the swapping arm. The molecular shape of full-length enzymes in solution reveals the localization and variation in flexibility of N- and C-terminal segments.
Organizational Affiliation:
Institute of Enzimology, BRC, Hungarian Academy of Sciences, Budapest, Hungary.