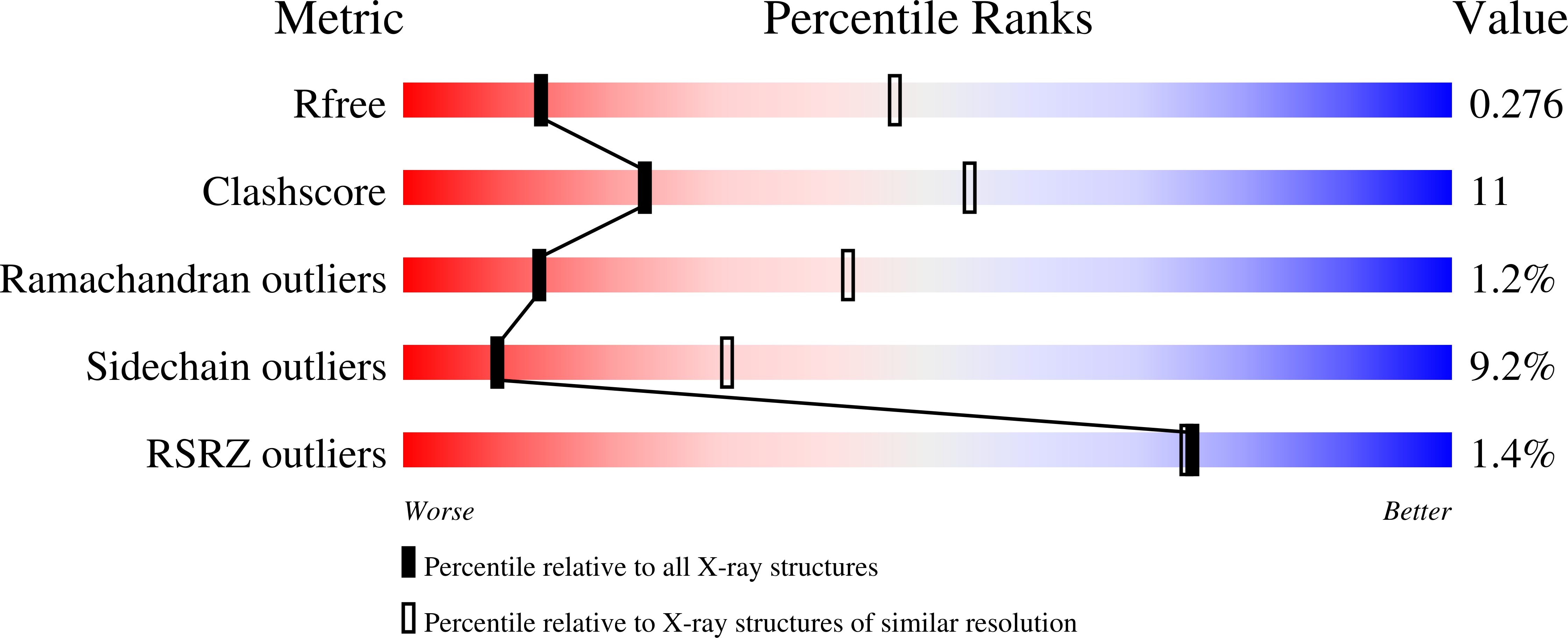
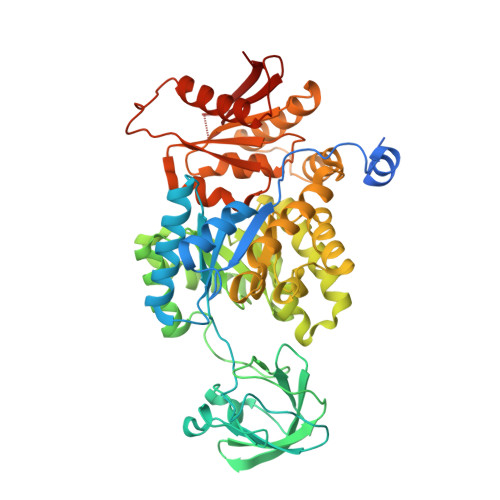
Sulphate removal induces a major conformational change in Leishmania mexicana pyruvate kinase in the crystalline state.
Tulloch, L.B., Morgan, H.P., Hannaert, V., Michels, P.A., Fothergill-Gilmore, L.A., Walkinshaw, M.D.(2008) J Mol Biol 383: 615-626
- PubMed: 18775437
- DOI: https://doi.org/10.1016/j.jmb.2008.08.037
- Primary Citation of Related Structures:
3E0V, 3E0W - PubMed Abstract:
We report X-ray structures of pyruvate kinase from Leishmania mexicana (LmPYK) that are trapped in different conformations. These, together with the previously reported structure of LmPYK in its inactive (T-state) conformation, allow comparisons of three different conformers of the same species of pyruvate kinase (PYK). Four new site point mutants showing the effects of side-chain alteration at subunit interfaces are also enzymatically characterised. The LmPYK tetramer crystals grown with ammonium sulphate as precipitant adopt an active-like conformation, with sulphate ions at the active and effector sites. The sulphates occupy positions similar to those of the phosphates of ligands bound to active (R-state) and constitutively active (nonallosteric) PYKs from several species, and provide insight into the structural roles of the phosphates of the substrates and effectors. Crystal soaking in sulphate-free buffers was found to induce major conformational changes in the tetramer. In particular, the unwinding of the Aalpha6' helix and the inward hinge movement of the B domain are coupled with a significant widening (4 A) of the tetramer caused by lateral movement of the C domains. The two new LmPYK structures and the activity studies of site point mutations described in this article are consistent with a developing picture of allosteric activity in which localised changes in protein flexibility govern the distribution of conformer families adopted by the tetramer in its active and inactive states.
Organizational Affiliation:
Institute of Structural and Molecular Biology, The University of Edinburgh, Michael Swann Building, The King's Buildings, Edinburgh, UK.