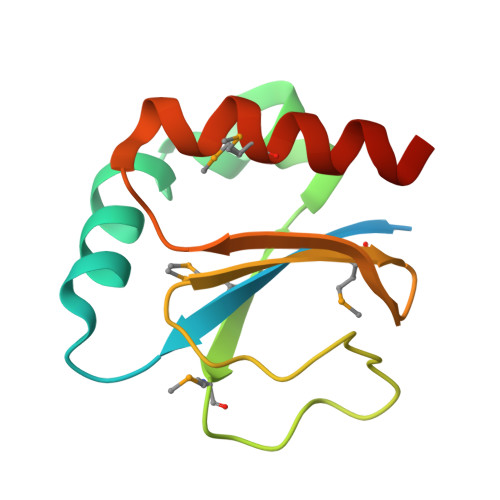
Crystal structure of the periplasmic thioredoxin SoxS from Paracoccus pantotrophus indicates a dual Trx/Grx functionality for activation of chemotrophic sulfur oxidation in vivo
Carius, Y., Rother, D., Friedrich, C.G., Scheidig, A.J.To be published.