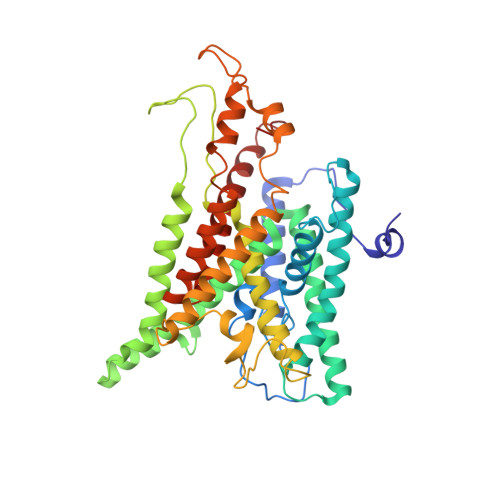
Single copies of Sec61 and TRAP associate with a nontranslating mammalian ribosome.
Menetret, J.F., Hegde, R.S., Aguiar, M., Gygi, S.P., Park, E., Rapoport, T.A., Akey, C.W.(2008) Structure 16: 1126-1137
- PubMed: 18611385
- DOI: https://doi.org/10.1016/j.str.2008.05.003
- Primary Citation of Related Structures:
3DKN - PubMed Abstract:
During cotranslational protein translocation, the ribosome associates with a membrane channel, formed by the Sec61 complex, and recruits the translocon-associated protein complex (TRAP). Here we report the structure of a ribosome-channel complex from mammalian endoplasmic reticulum in which the channel has been visualized at 11 A resolution. In this complex, single copies of Sec61 and TRAP associate with a nontranslating ribosome and this stoichiometry was verified by quantitative mass spectrometry. A bilayer-like density surrounds the channel and can be attributed to lipid and detergent. The crystal structure of an archaeal homolog of the Sec61 complex was then docked into the map. In this model, two cytoplasmic loops of Sec61 may interact with RNA helices H6, H7, and H50, while the central pore is located below the ribosome tunnel exit. Hence, this copy of Sec61 is positioned to capture and translocate the nascent chain. Finally, we show that mammalian and bacterial ribosome-channel complexes have similar architectures.
Organizational Affiliation:
Department of Physiology and Biophysics, Boston University School of Medicine, 700 Albany Street, Boston, MA 02118-2526, USA.