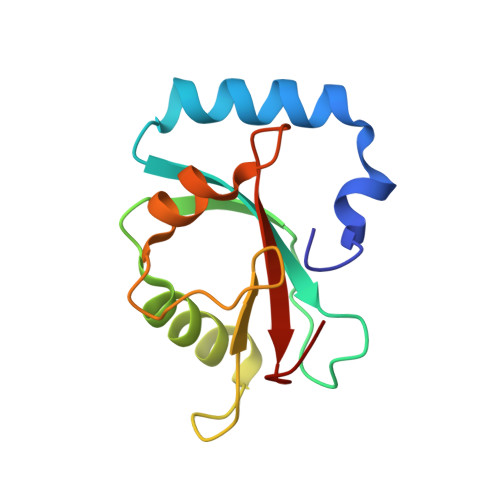
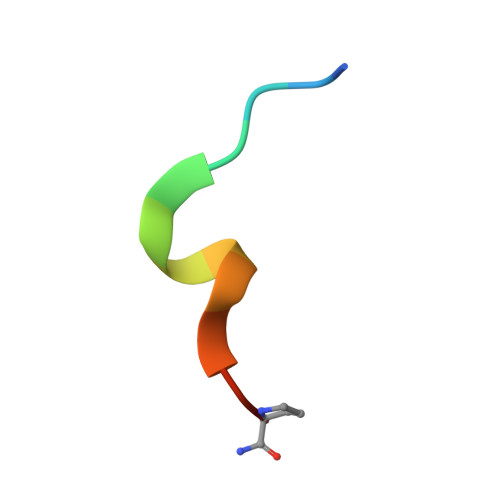
Ligand Binding Mode of GABA(A) Receptor-Associated Protein.
Weiergraber, O.H., Stangler, T., Thielmann, Y., Mohrluder, J., Wiesehan, K., Willbold, D.(2008) J Mol Biol 381: 1320-1331
- PubMed: 18638487
- DOI: https://doi.org/10.1016/j.jmb.2008.06.086
- Primary Citation of Related Structures:
3D32 - PubMed Abstract:
The gamma-aminobutyric acid type A (GABA(A)) receptor-associated protein is a versatile adaptor protein playing an important role in intracellular vesicle trafficking, particularly in neuronal cells. We present the X-ray structure of the soluble form of human GABA(A) receptor-associated protein complexed with a high-affinity synthetic peptide at 1.3 A resolution. The data shed light on the probable binding modes of key interaction partners, including the GABA(A) receptor and the cysteine protease Atg4. The resulting models provide a structural background for further investigation of the unique biological properties of this protein.
Organizational Affiliation:
Institut für Neurowissenschaften und Biophysik, Molekulare Biophysik, Forschungszentrum Jülich, D-52425 Jülich, Germany.