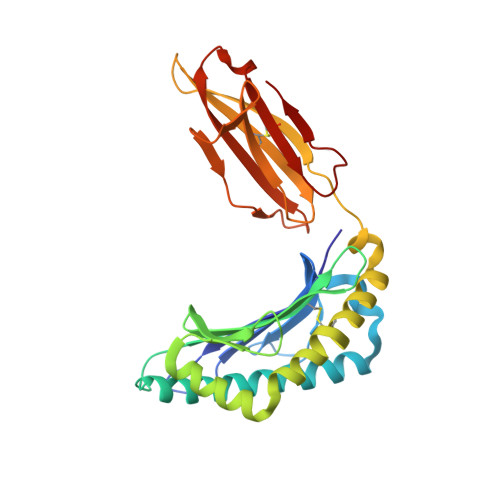
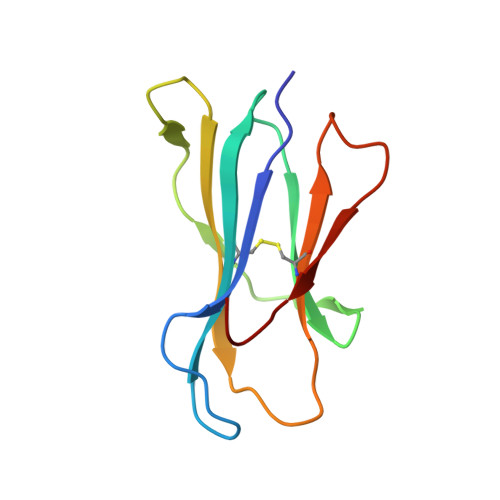
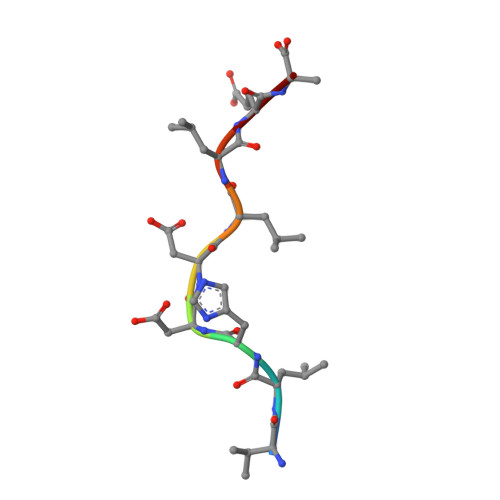
Secondary anchor polymorphism in the HA-1 minor histocompatibility antigen critically affects MHC stability and TCR recognition
Nicholls, S., Piper, K.P., Mohammed, F., Dafforn, T.R., Tenzer, S., Salim, M., Mahendra, P., Craddock, C., van Endert, P., Schild, H., Cobbold, M., Engelhard, V.H., Moss, P.A., Willcox, B.E.(2009) Proc Natl Acad Sci U S A 106: 3889-3894
- PubMed: 19234124
- DOI: https://doi.org/10.1073/pnas.0900411106
- Primary Citation of Related Structures:
3D25 - PubMed Abstract:
T cell recognition of minor histocompatibility antigens (mHags) underlies allogeneic immune responses that mediate graft-versus-host disease and the graft-versus-leukemia effect following stem cell transplantation. Many mHags derive from single amino acid polymorphisms in MHC-restricted epitopes, but our understanding of the molecular mechanisms governing mHag immunogenicity and recognition is incomplete. Here we examined antigenic presentation and T-cell recognition of HA-1, a prototypic autosomal mHag derived from single nucleotide dimorphism (HA-1(H) versus HA-1(R)) in the HMHA1 gene. The HA-1(H) peptide is restricted by HLA-A2 and is immunogenic in HA-1(R/R) into HA-1(H) transplants, while HA-1(R) has been suggested to be a "null allele" in terms of T cell reactivity. We found that proteasomal cleavage and TAP transport of the 2 peptides is similar and that both variants can bind to MHC. However, the His>Arg change substantially decreases the stability and affinity of HLA-A2 association, consistent with the reduced immunogenicity of the HA-1(R) variant. To understand these findings, we determined the structure of an HLA-A2-HA-1(H) complex to 1.3A resolution. Whereas His-3 is accommodated comfortably in the D pocket, incorporation of the lengthy Arg-3 is predicted to require local conformational changes. Moreover, a soluble TCR generated from HA-1(H)-specific T-cells bound HA-1(H) peptide with moderate affinity but failed to bind HA-1(R), indicating complete discrimination of HA-1 variants at the level of TCR/MHC interaction. Our results define the molecular mechanisms governing immunogenicity of HA-1, and highlight how single amino acid polymorphisms in mHags can critically affect both MHC association and TCR recognition.
Organizational Affiliation:
Cancer Research UK Institute for Cancer Studies, Edgbaston, Birmingham B15 2TT, United Kingdom.