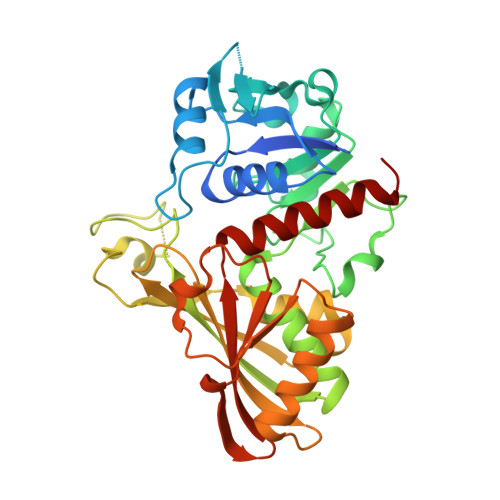
Crystal structure of Cryptosporidium parvum glyceraldehyde-3-phosphate dehydrogenase.
Wernimont, A.K., Lew, J., Kozieradzki, I., Cossar, D., Schapiro, M., Bochkarev, A., Bountra, C., Weigelt, J., Edwards, A.M., Hui, R., Pizarro, J., Hills, T.To be published.