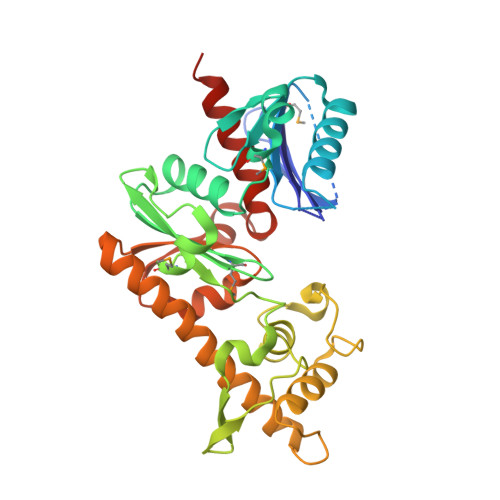
Crystal structure of the pantheonate kinase-like protein Q6M145 at the resolution 1.8 A. Northeast Structural Genomics Consortium target MrR63.
Kuzin, A.P., Su, M., Seetharaman, J., Forouhar, F., Wang, D., Fang, Y., Cunningham, K., Ma, L.-C., Xiao, R., Liu, J., Baran, J.F., Acton, T.B., Rost, B., Montelione, G.T., Hunt, J.F., Tong, L.To be published.