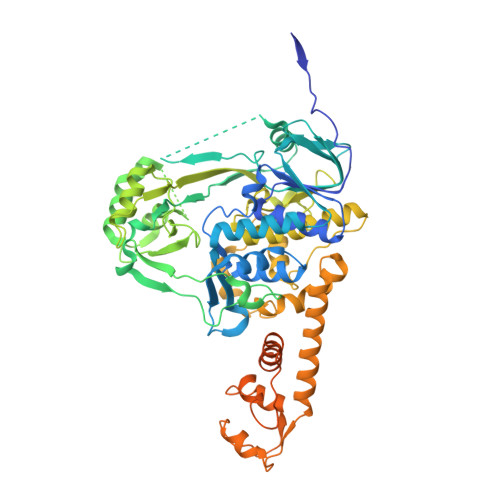
Structure-function analysis of Escherichia coli MnmG (GidA), a highly conserved tRNA-modifying enzyme.
Shi, R., Villarroya, M., Ruiz-Partida, R., Li, Y., Proteau, A., Prado, S., Moukadiri, I., Benitez-Paez, A., Lomas, R., Wagner, J., Matte, A., Velazquez-Campoy, A., Armengod, M.E., Cygler, M.(2009) J Bacteriol 191: 7614-7619
- PubMed: 19801413
- DOI: https://doi.org/10.1128/JB.00650-09
- Primary Citation of Related Structures:
3CES, 3G05 - PubMed Abstract:
The MnmE-MnmG complex is involved in tRNA modification. We have determined the crystal structure of Escherichia coli MnmG at 2.4-A resolution, mutated highly conserved residues with putative roles in flavin adenine dinucleotide (FAD) or tRNA binding and MnmE interaction, and analyzed the effects of these mutations in vivo and in vitro. Limited trypsinolysis of MnmG suggests significant conformational changes upon FAD binding.
Organizational Affiliation:
Department of Biochemistry, McGill University, Montreal, Quebec, Canada.