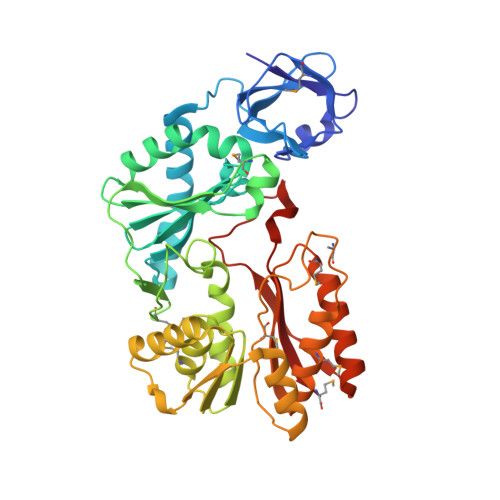
Crystal structure of the Escherichia coli 23S rRNA:m5C methyltransferase RlmI (YccW) reveals evolutionary links between RNA modification enzymes
Sunita, S., Tkaczuk, K.L., Purta, E., Kasprzak, J.M., Douthwaite, S., Bujnicki, J.M., Sivaraman, J.(2008) J Mol Biol 383: 652-666
- PubMed: 18789337
- DOI: https://doi.org/10.1016/j.jmb.2008.08.062
- Primary Citation of Related Structures:
3C0K - PubMed Abstract:
Methylation is the most common RNA modification in the three domains of life. Transfer of the methyl group from S-adenosyl-l-methionine (AdoMet) to specific atoms of RNA nucleotides is catalyzed by methyltransferase (MTase) enzymes. The rRNA MTase RlmI (rRNA large subunit methyltransferase gene I; previously known as YccW) specifically modifies Escherichia coli 23S rRNA at nucleotide C1962 to form 5-methylcytosine. Here, we report the crystal structure of RlmI refined at 2 A to a final R-factor of 0.194 (R(free)=0.242). The RlmI molecule comprises three domains: the N-terminal PUA domain; the central domain, which resembles a domain previously found in RNA:5-methyluridine MTases; and the C-terminal catalytic domain, which contains the AdoMet-binding site. The central and C-terminal domains are linked by a beta-hairpin structure that has previously been observed in several MTases acting on nucleic acids or proteins. Based on bioinformatics analyses, we propose a model for the RlmI-AdoMet-RNA complex. Comparative structural analyses of RlmI and its homologs provide insight into the potential function of several structures that have been solved by structural genomics groups and furthermore indicate that the evolutionary paths of RNA and DNA 5-methyluridine and 5-methylcytosine MTases have been closely intertwined.
Organizational Affiliation:
Department of Biological Sciences, National University of Singapore, Singapore.