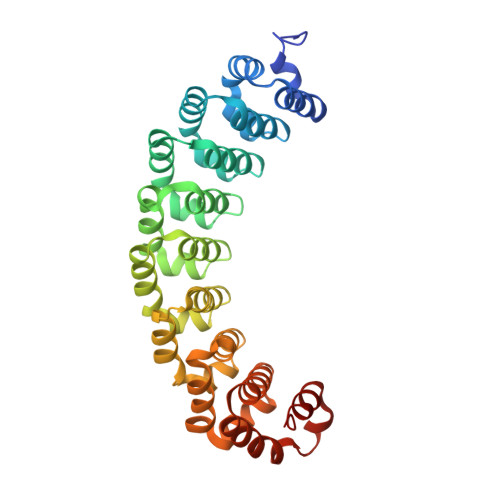
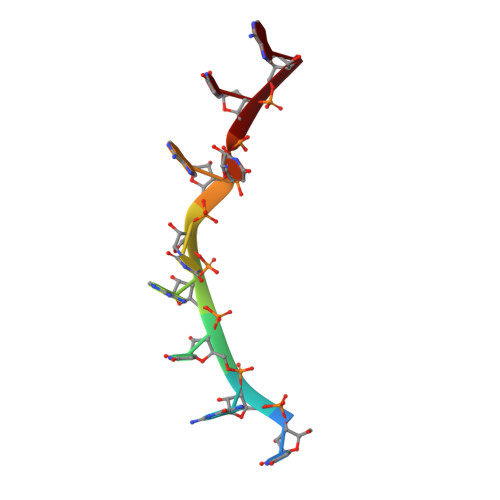
Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p
Miller, M.T., Higgin, J.J., Hall, T.M.T.(2008) Nat Struct Mol Biol 15: 397-402
- PubMed: 18327269
- DOI: https://doi.org/10.1038/nsmb.1390
- Primary Citation of Related Structures:
3BWT, 3BX2, 3BX3 - PubMed Abstract:
Pumilio/FBF (PUF) family proteins are found in eukaryotic organisms and regulate gene expression post-transcriptionally by binding to sequences in the 3' untranslated region of target transcripts. PUF proteins contain an RNA binding domain that typically comprises eight alpha-helical repeats, each of which recognizes one RNA base. Some PUF proteins, including yeast Puf4p, have altered RNA binding specificity and use their eight repeats to bind to RNA sequences with nine or ten bases. Here we report the crystal structures of Puf4p alone and in complex with a 9-nucleotide (nt) target RNA sequence, revealing that Puf4p accommodates an 'extra' nucleotide by modest adaptations allowing one base to be turned away from the RNA binding surface. Using structural information and sequence comparisons, we created a mutant Puf4p protein that preferentially binds to an 8-nt target RNA sequence over a 9-nt sequence and restores binding of each protein repeat to one RNA base.
Organizational Affiliation:
Laboratory of Structural Biology, National Institute of Environmental Health Sciences, National Institutes of Health, 111 T.W. Alexander Drive, Building 101, Room F363, Research Triangle Park, North Carolina 27709, USA.