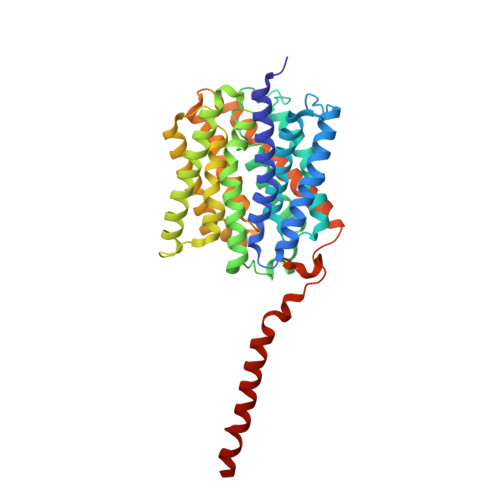
Structure of the Nitrosomonas europaea Rh protein.
Li, X., Jayachandran, S., Nguyen, H.H., Chan, M.K.(2007) Proc Natl Acad Sci U S A 104: 19279-19284
- PubMed: 18040042
- DOI: https://doi.org/10.1073/pnas.0709710104
- Primary Citation of Related Structures:
3B9Y, 3B9Z - PubMed Abstract:
Amt/MEP/Rh proteins are a family of integral membrane proteins implicated in the transport of NH3, CH(2)NH2, and CO2. Whereas Amt/MEP proteins are agreed to transport ammonia (NH3/NH4+), the primary substrate for Rh proteins has been controversial. Initial studies suggested that Rh proteins also transport ammonia, but more recent evidence suggests that they transport CO2. Here we report the first structure of an Rh family member, the Rh protein from the chemolithoautotrophic ammonia-oxidizing bacterium Nitrosomonas europaea. This Rh protein exhibits a number of similarities to its Amt cousins, including a trimeric oligomeric state, a central pore with an unusual twin-His site in the middle, and a Phe residue that blocks the channel for small-molecule transport. However, there are some significant differences, the most notable being the presence of an additional cytoplasmic C-terminal alpha-helix, an increased number of internal proline residues along the transmembrane helices, and a specific set of residues that appear to link the C-terminal helix to Phe blockage. This latter linkage suggests a mechanism in which binding of a partner protein to the C terminus could regulate channel opening. Another difference is the absence of the extracellular pi-cation binding site conserved in Amt/Mep structures. Instead, CO2 pressurization experiments identify a CO2 binding site near the intracellular exit of the channel whose residues are highly conserved in all Rh proteins, except those belonging to the Rh30 subfamily. The implications of these findings on the functional role of the human Rh antigens are discussed.
Organizational Affiliation:
Biophysics Program and Departments of Biochemistry and Chemistry, Ohio State University, 484 West Twelfth Avenue, Columbus, OH 43210, USA.