The triple helical structure and stability of collagen model peptide with 4(S)-hydroxyprolyl-pro-gly units
Motooka, D., Kawahara, K., Nakamura, S., Doi, M., Nishi, Y., Nishiuchi, Y., Kee Kang, Y., Nakazawa, T., Uchiyama, S., Yoshida, T., Ohkubo, T., Kobayashi, Y.(2011) Biopolymers 98: 111-121
- PubMed: 22020801
- DOI: https://doi.org/10.1002/bip.21730
- Primary Citation of Related Structures:
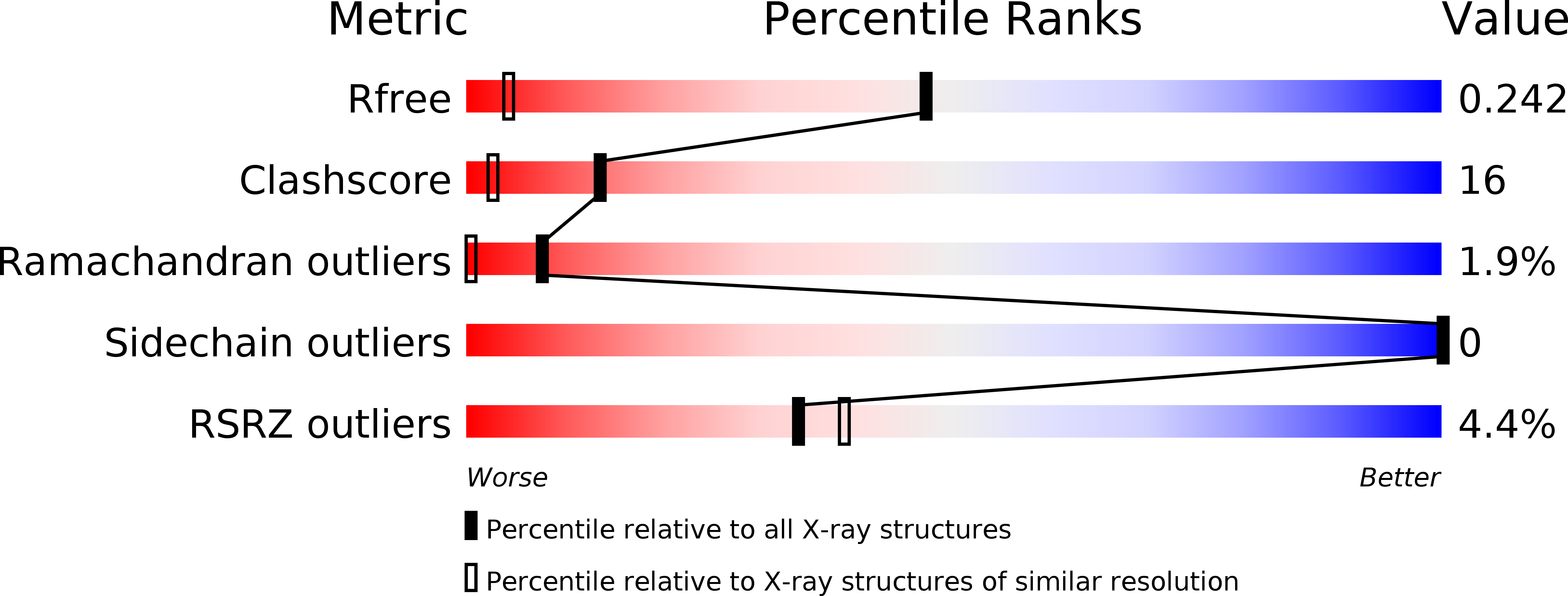
3B2C - PubMed Abstract:
Extensive studies on the structure of collagen have revealed that the hydroxylation of Pro residues in a variety of model peptides with the typical (X-Y-Gly)(n) repeats (X and Y: Pro and its analogues) represents one of the major factors influencing the stability of triple helices. While(2S,4R)-hydroxyproline (Hyp) at the position Y stabilizes the triple helix, (2S,4S)-hydroxyproline (hyp) at the X-position destabilizes the helix as demonstrated that the triple helix of (hyp-Pro-Gly)(15) is less stable than that of (Pro-Pro-Gly)(15) and that a shorter peptide (hyp-Pro-Gly)(10) does not form the helix. To clarify the role of the hydroxyl group of Pro residues to play in the stabilization mechanism of the collagen triple helix, we synthesized and crystallized a model peptide (Pro-Hyp-Gly)(4) -(hyp-Pro-Gly)(2) -(Pro-Hyp-Gly)(4) and analyzed its structure by X-ray crystallography and CD spectroscopy. In the crystal, the main-chain of this peptide forms a typical collagen like triple helix. The majority of hyp residues take down pucker with exceptionally shallow angles probably to relieve steric hindrance, but the remainders protrude the hydroxyl group toward solvent with the less favorable up pucker to fit in a triple helix. There is no indication of the existence of an intra-molecular hydrogen bond between the hydroxyl moiety and the carbonyl oxygen of hyp supposed to destabilize the triple helix. We also compared the conformational energies of up and down packers of the pyrrolidine ring in Ac-hyp-NMe(2) by quantum mechanical calculations.
Organizational Affiliation:
Graduate School of Pharmaceutical Sciences, Osaka University, Osaka, Japan.