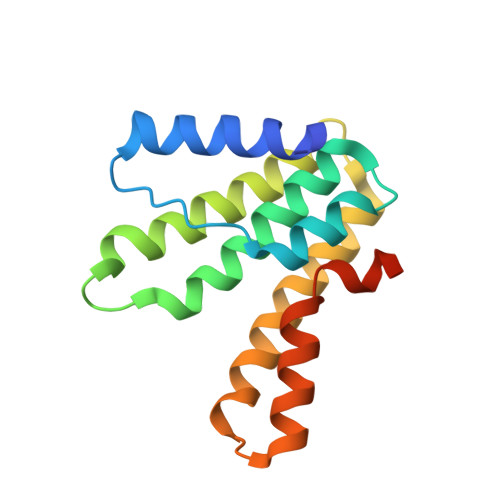
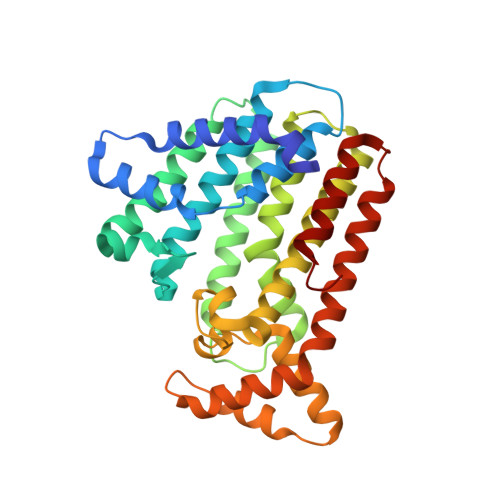
Crystal structure of heterodimeric hexaprenyl diphosphate synthase from Micrococcus luteus B-P 26 reveals that the small subunit is directly involved in the product chain length regulation.
Sasaki, D., Fujihashi, M., Okuyama, N., Kobayashi, Y., Noike, M., Koyama, T., Miki, K.(2011) J Biol Chem 286: 3729-3740
- PubMed: 21068379
- DOI: https://doi.org/10.1074/jbc.M110.147991
- Primary Citation of Related Structures:
3AQB, 3AQC - PubMed Abstract:
Hexaprenyl diphosphate synthase from Micrococcus luteus B-P 26 (Ml-HexPPs) is a heterooligomeric type trans-prenyltransferase catalyzing consecutive head-to-tail condensations of three molecules of isopentenyl diphosphates (C(5)) on a farnesyl diphosphate (FPP; C(15)) to form an (all-E) hexaprenyl diphosphate (HexPP; C(30)). Ml-HexPPs is known to function as a heterodimer of two different subunits, small and large subunits called HexA and HexB, respectively. Compared with homooligomeric trans-prenyltransferases, the molecular mechanism of heterooligomeric trans-prenyltransferases is not yet clearly understood, particularly with respect to the role of the small subunits lacking the catalytic motifs conserved in most known trans-prenyltransferases. We have determined the crystal structure of Ml-HexPPs both in the substrate-free form and in complex with 7,11-dimethyl-2,6,10-dodecatrien-1-yl diphosphate ammonium salt (3-DesMe-FPP), an analog of FPP. The structure of HexB is composed of mostly antiparallel α-helices joined by connecting loops. Two aspartate-rich motifs (designated the first and second aspartate-rich motifs) and the other characteristic motifs in HexB are located around the diphosphate part of 3-DesMe-FPP. Despite the very low amino acid sequence identity and the distinct polypeptide chain lengths between HexA and HexB, the structure of HexA is quite similar to that of HexB. The aliphatic tail of 3-DesMe-FPP is accommodated in a large hydrophobic cleft starting from HexB and penetrating to the inside of HexA. These structural features suggest that HexB catalyzes the condensation reactions and that HexA is directly involved in the product chain length control in cooperation with HexB.
Organizational Affiliation:
Department of Chemistry, Graduate School of Science, Kyoto University, Sakyo-ku, Kyoto, Kyoto 606-8502, Japan.