A novel intein-like autoproteolytic mechanism in autotransporter proteins.
Tajima, N., Kawai, F., Park, S.Y., Tame, J.R.(2010) J Mol Biol 402: 645-656
- PubMed: 20615416
- DOI: https://doi.org/10.1016/j.jmb.2010.06.068
- Primary Citation of Related Structures:
3AEH - PubMed Abstract:
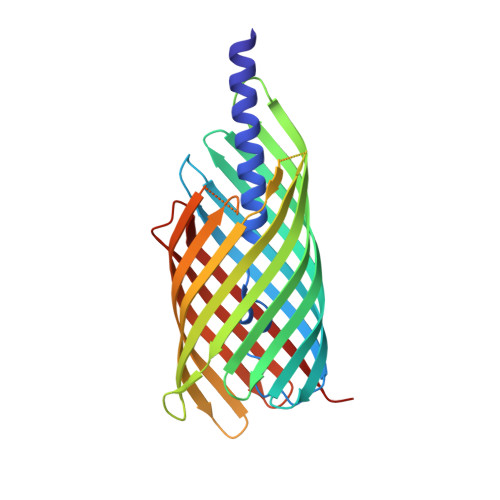
Many virulence factors secreted by pathogenic Gram-negative bacteria are found to be members of the autotransporter protein family. These proteins share a common mechanism by which they exit the periplasm, involving the formation of a 12-stranded β-barrel domain in the outer membrane. The role of this barrel in the secretion of the N-terminal passenger domain is controversial, and no model currently explains satisfactorily the entire body of experimental data. After secretion, some autotransporter barrels autoproteolytically cleave away the passenger, and one crystal structure is known for a barrel of this type in the postcleavage state. Hbp is an autotransporter of the self-cleaving type, which cuts the polypeptide between two absolutely conserved asparagine residues buried within the barrel lumen. Mutation of the first asparagine residue to isosteric aspartic acid prevents proteolysis. Here we present the crystal structure of a truncated Hbp mutant carrying the C-terminal residues of the passenger domain attached to the barrel. This model mimics the state of the protein immediately prior to separation of the passenger and barrel domains, and shows the role of residues in the so-called "linker" between the passenger and β domains. This high-resolution membrane protein crystal structure also reveals the sites of many water molecules within the barrel. The cleavage mechanism shows similarities to those of inteins and some viral proteins, but with a novel means of promoting nucleophilic attack.
Organizational Affiliation:
Yokohama City University, Suehiro 1-7-29, Tsurumi, Yokohama 230-0045, Japan.