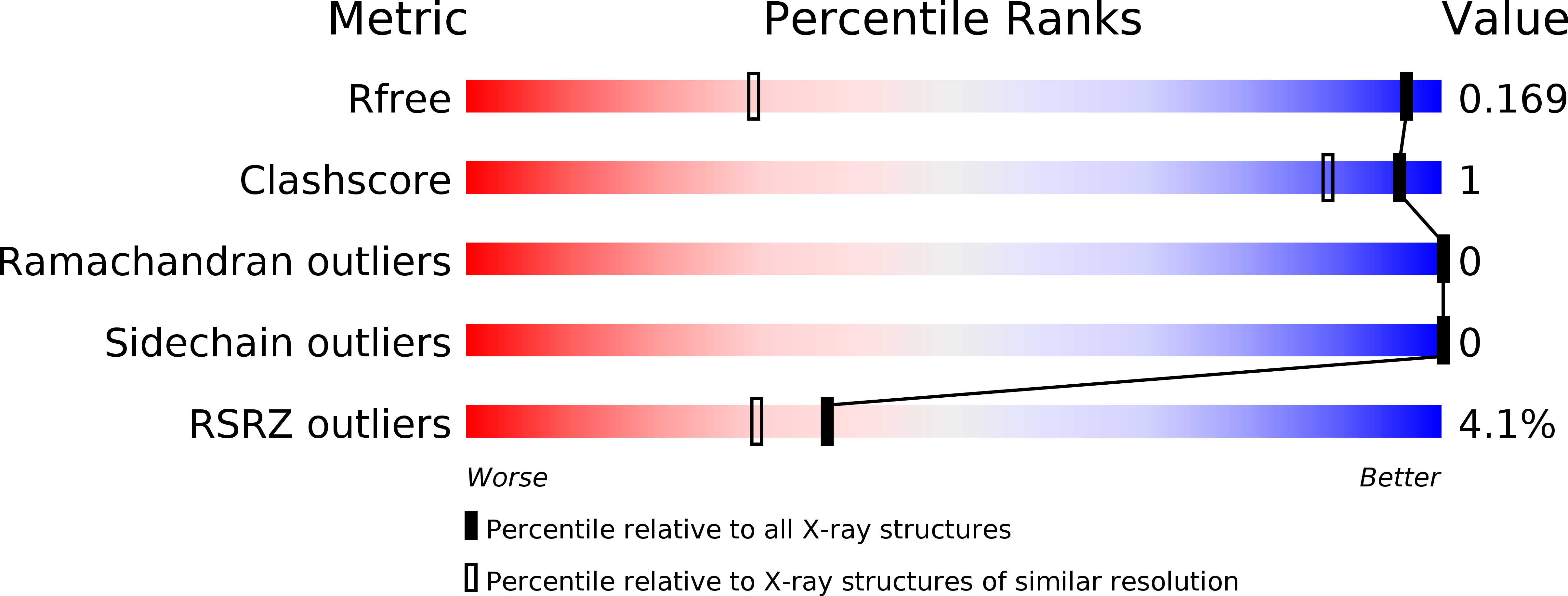
Crystal structure of the collagen model peptide (Pro-Pro-Gly)4 -Hyp-Asp-Gly-(Pro-Pro-Gly)4 at 1.0 angstrom resolution.
Okuyama, K., Kawaguchi, T., Shimura, M., Noguchi, K., Mizuno, K., Bachinger, H.P.(2013) Biopolymers 99: 436-447
- PubMed: 23616212
- DOI: https://doi.org/10.1002/bip.22198
- Primary Citation of Related Structures:
3ABN - PubMed Abstract:
The single-crystal structure of the collagen-like peptide (Pro-Pro-Gly)4 -Hyp-Asp-Gly-(Pro-Pro-Gly)4, was analyzed at 1.02 Å resolution. The overall average helical twist (θ = 49.6°) suggests that this peptide adopts a 7/2 triple-helical structure and that its conformation is very similar to that of (Gly-Pro-Hyp)9, which has the typical repeating sequence in collagen. High-resolution studies on other collagen-like peptides have shown that imino acid-rich sequences preferentially adopt a 7/2 triple-helical structure (θ = 51.4°), whereas imino acid-lean sequences adopt relaxed conformations (θ < 51.4°). The guest Gly-Hyp-Asp sequence in the present peptide, however, has a large helical twist (θ = 61.1°), whereas that of the host Pro-Pro-Gly sequence is small (θ = 46.7°), indicating that the relationship between the helical conformation and the amino acid sequence of such peptides is complex. In the present structure, a strong intermolecular hydrogen bond between two Asp residues on the A and B strands might induce the large helical twist of the guest sequence; this is compensated by a reduced helical twist in the host, so that an overall 7/2-helical symmetry is maintained. The Asp residue in the C strand might interact electrostatically with the N-terminus of an adjacent molecule, causing axial displacement, reminiscent of the D-staggered structure in fibrous collagens.
Organizational Affiliation:
Department of Macromolecular Science, Graduate School of Science, Osaka University, Toyonaka, Osaka, 560-0043, Japan. okuyamak@chem.sci.osaka-u.ac.jp