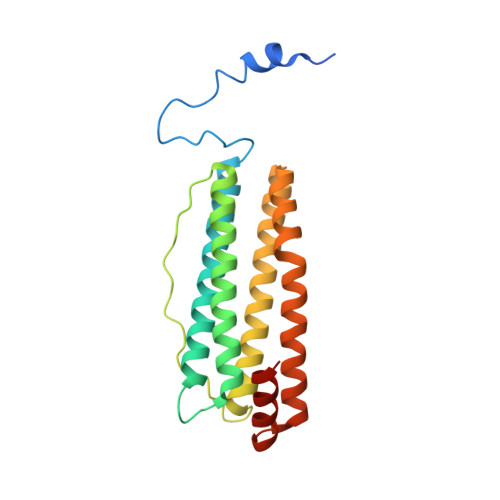
Crystal structure of plant ferritin reveals a novel metal binding site that functions as a transit site for metal transfer in ferritin
Masuda, T., Goto, F., Yoshihara, T., Mikami, B.(2010) J Biol Chem 285: 4049-4059
- PubMed: 20007325
- DOI: https://doi.org/10.1074/jbc.M109.059790
- Primary Citation of Related Structures:
3A68, 3A9Q - PubMed Abstract:
Ferritins are important iron storage and detoxification proteins that are widely distributed in living kingdoms. Because plant ferritin possesses both a ferroxidase site and a ferrihydrite nucleation site, it is a suitable model for studying the mechanism of iron storage in ferritin. This article presents for the first time the crystal structure of a plant ferritin from soybean at 1.8-A resolution. The soybean ferritin 4 (SFER4) had a high structural similarity to vertebrate ferritin, except for the N-terminal extension region, the C-terminal short helix E, and the end of the BC-loop. Similar to the crystal structures of other ferritins, metal binding sites were observed in the iron entry channel, ferroxidase center, and nucleation site of SFER4. In addition to these conventional sites, a novel metal binding site was discovered intermediate between the iron entry channel and the ferroxidase site. This site was coordinated by the acidic side chain of Glu(173) and carbonyl oxygen of Thr(168), which correspond, respectively, to Glu(140) and Thr(135) of human H chain ferritin according to their sequences. A comparison of the ferroxidase activities of the native and the E173A mutant of SFER4 clearly showed a delay in the iron oxidation rate of the mutant. This indicated that the glutamate residue functions as a transit site of iron from the 3-fold entry channel to the ferroxidase site, which may be universal among ferritins.
Organizational Affiliation:
From the Laboratory of Food Quality Design and Development, Division of Agronomy and Horticultural Science, Graduate School of Agriculture, Kyoto University, Gokasho, Uji, Kyoto 611-0011. Electronic address: masutaro@kais.kyoto-u.ac.jp.