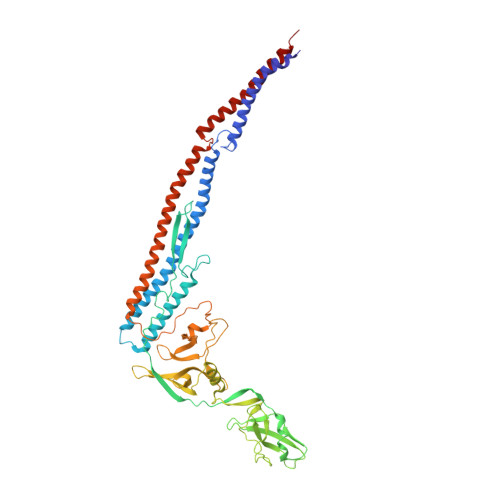
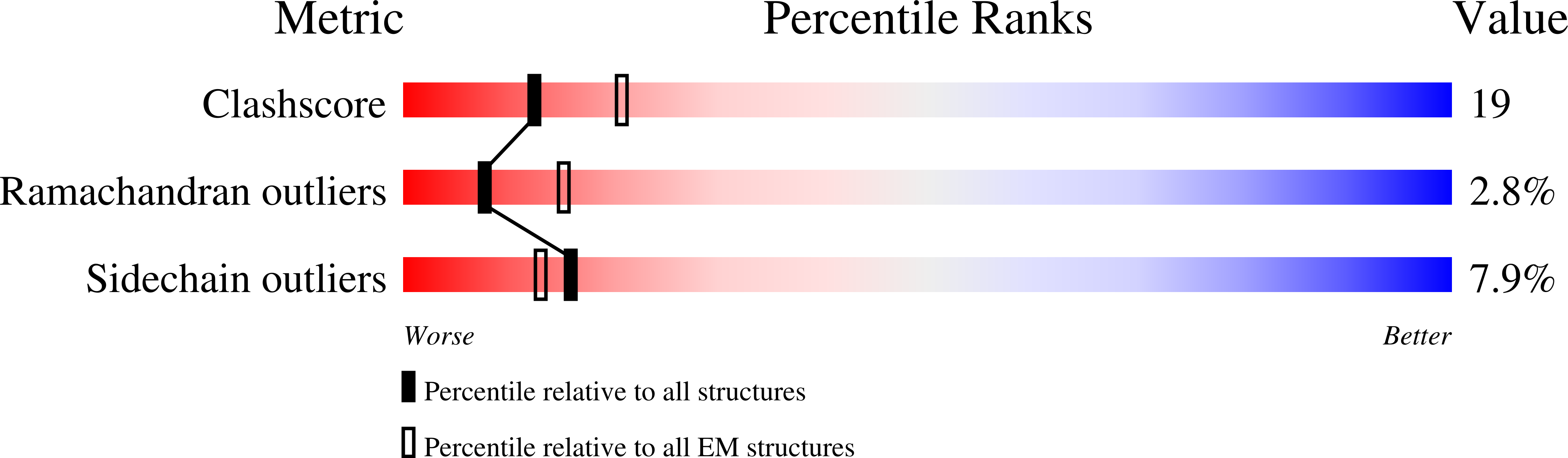Conformational change of flagellin for polymorphic supercoiling of the flagellar filament
Maki-Yonekura, S., Yonekura, K., Namba, K.(2010) Nat Struct Mol Biol 17: 417-422
- PubMed: 20228803
- DOI: https://doi.org/10.1038/nsmb.1774
- Primary Citation of Related Structures:
3A5X - PubMed Abstract:
The bacterial flagellar filament is a helical propeller rotated by the flagellar motor for bacterial locomotion. The filament is a supercoiled assembly of a single protein, flagellin, and is formed by 11 protofilaments. For bacterial taxis, the reversal of motor rotation switches the supercoil between left- and right-handed, both of which arise from combinations of two distinct conformations and packing interactions of the L-type and R-type protofilaments. Here we report an atomic model of the L-type straight filament by electron cryomicroscopy and helical image analysis. Comparison with the R-type structure shows interesting features: an orientation change of the outer core domains (D1) against the inner core domains (D0) showing almost invariant orientation and packing, a conformational switching within domain D1, and the conformational flexibility of domains D0 and D1 with their spoke-like connection for tight molecular packing.
Organizational Affiliation:
Dynamic NanoMachine Project, International Cooperative Research Project, Japan Science and Technology Agency, Suita, Osaka, Japan.