Structural Basis of Alternative DNA Recognition by Maf Transcription Factors
Kurokawa, H., Motohashi, H., Sueno, S., Kimura, M., Takagawa, H., Kanno, Y., Yamamoto, M., Tanaka, T.(2009) Mol Cell Biol 29: 6232-6244
- PubMed: 19797082
- DOI: https://doi.org/10.1128/MCB.00708-09
- Primary Citation of Related Structures:
3A5T - PubMed Abstract:
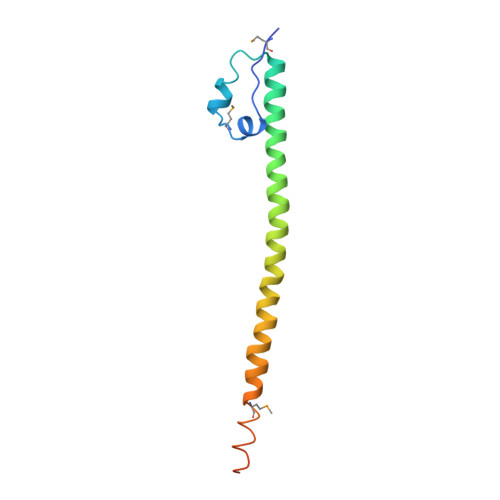
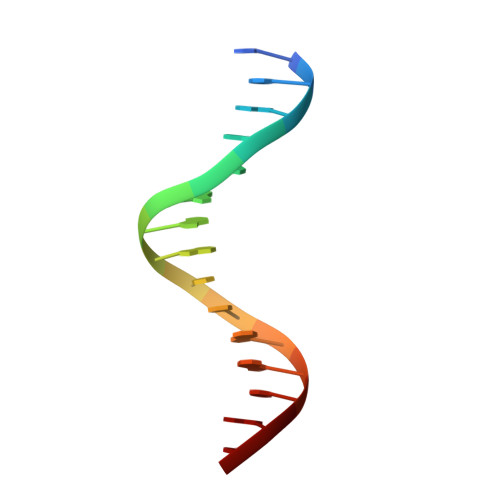
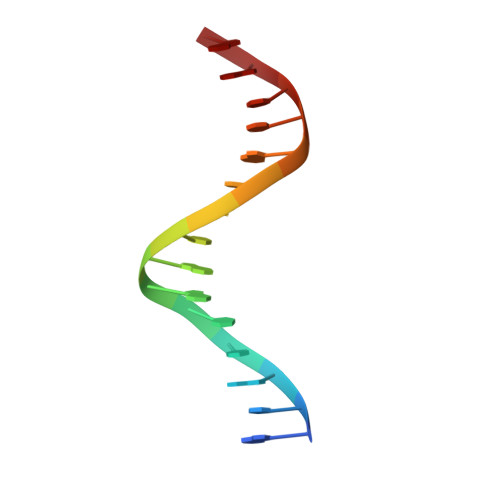
Maf transcription factors constitute a family of the basic region-leucine zipper (bZip) factors and recognize unusually long DNA motifs (13 or 14 bp), termed the Maf recognition element (MARE). The MARE harbors extended GC sequences on each side of its core motif, which is similar to TRE or CRE (7 or 8 bp) recognized by the AP1 and CREB/ATF families, respectively. To ascertain the structural basis governing the acquirement of such unique DNA recognition, we determined the crystal structure of the MafG-DNA complex. Each MafG monomer consists of three helices in which the carboxyl-terminal long helix organizes one DNA-contacting element and one coiled-coil dimer formation element. To our surprise, two well-conserved residues, Arg57 and Asn61 in the basic region, play critical roles in Maf-specific DNA recognition. These two residues show unique side-chain orientations and interact directly with the extended GC bases. Maf-specific residues in the amino-terminal and basic regions appear to indirectly stabilize MARE recognition through DNA backbone phosphate interactions. This study revealed an alternative DNA recognition mechanism of the bZip factors that bestows specific target gene profiles upon Maf homodimers or Maf-containing heterodimers.
Organizational Affiliation:
Institute of Multidisciplinary Research for Advanced Materials, Tohoku University, Sendai 980-8577, Japan. hkuro@m.tains.tohoku.ac.jp